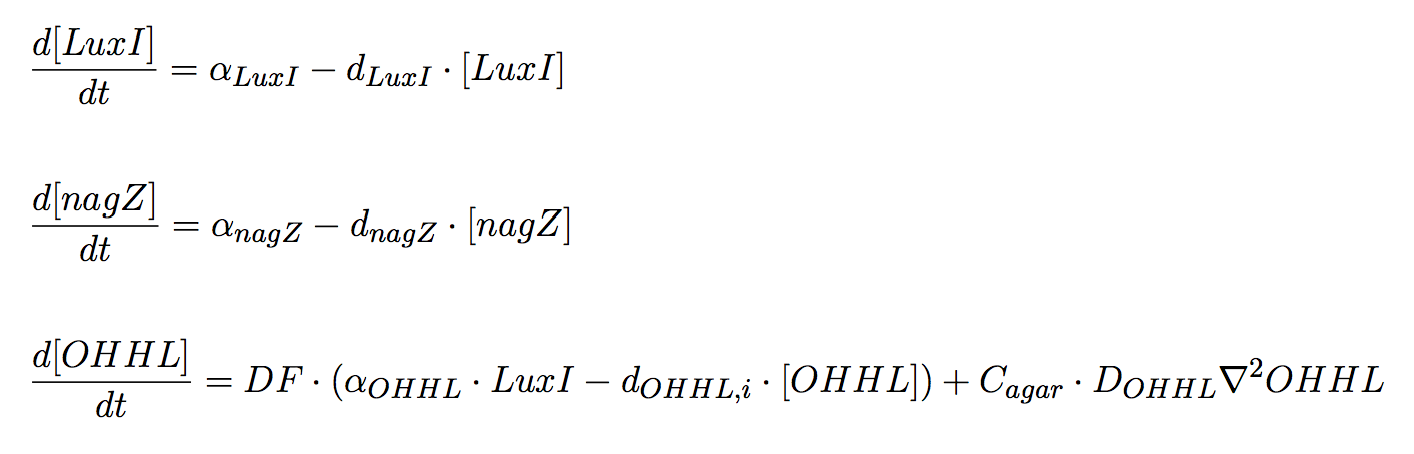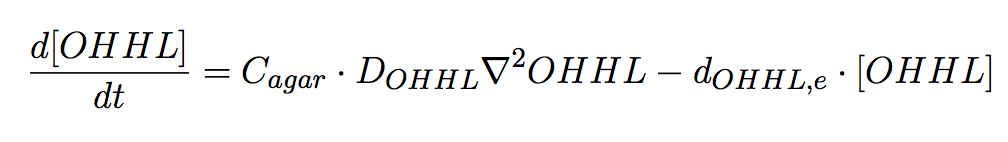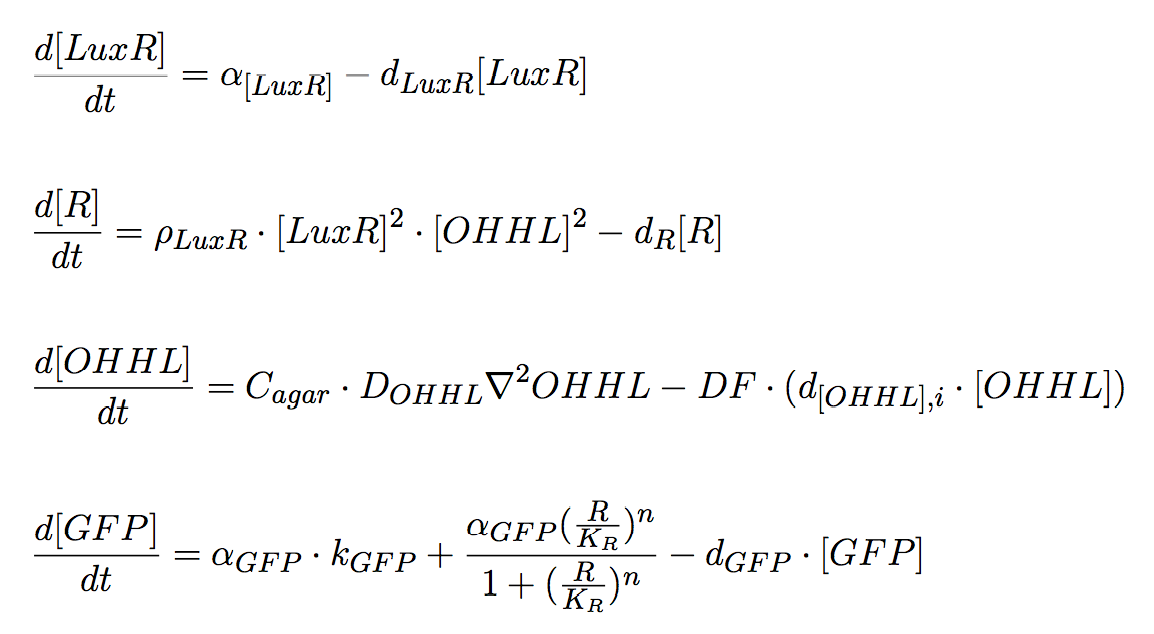Team:ETH Zurich/GFP
From 2013.igem.org
| Line 35: | Line 35: | ||
<h1>Receiver cells </h1> | <h1>Receiver cells </h1> | ||
| - | <p align="justify">Receiver cells are responsible for processing the signal sent by the mine cells. | + | <p align="justify">Receiver cells are responsible for processing the signal sent by the mine cells. OHHL acts as the input signal and the output is green fluorescence whose intensity correlates with the sensed OHHL concentration by receiver cells; for this purpose GFP expression is under control of pLux promoter. Once OHHL molecules reach a non-mine colony, it can bind to LuxR protein, and the complex LuxR/OHHL acts as a transcriptional activator. Therefore, this module works as a high pass amplitude filter for GFP, since it is produced when the OHHL levels are sufficiently high. <br><br> </p> |
| - | The | + | The system of equations for the states involved in the sender module are given below: |
[[File:receiver_GFP.png|500px|center|thumb|<b>Equation system 7: </b> System of Differential equations for receiver cells]] | [[File:receiver_GFP.png|500px|center|thumb|<b>Equation system 7: </b> System of Differential equations for receiver cells]] | ||
Revision as of 17:57, 2 October 2013
Contents |
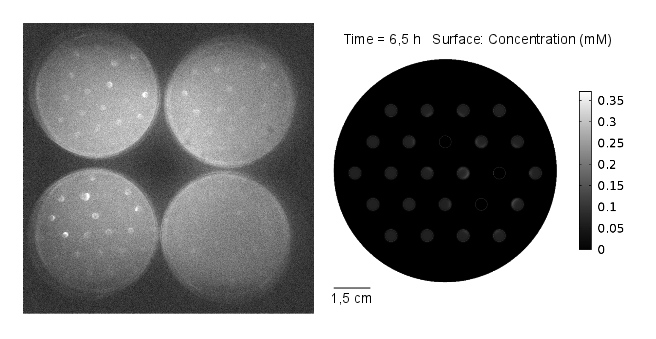
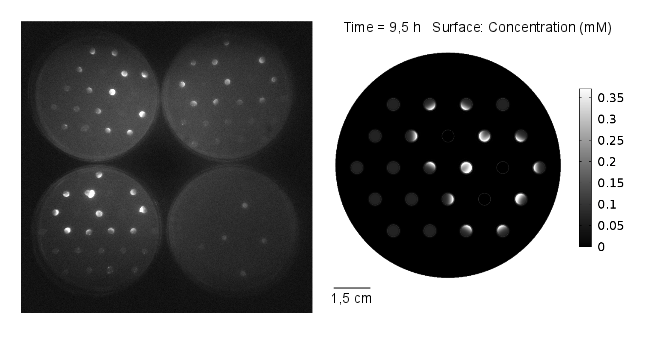
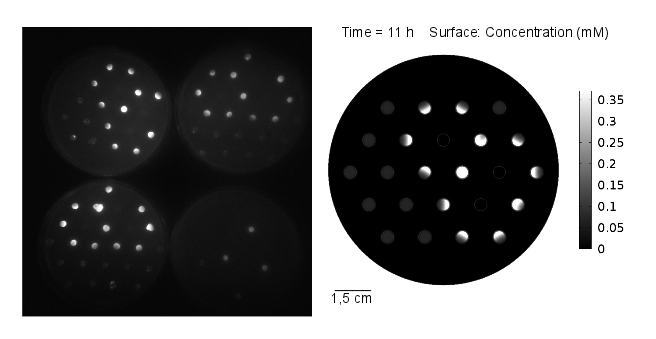
Reaction-diffusion model: genetic circuit with GFP as reporter gene
In our first spatio-temporal model, we wanted to find out if (i) a suitable OHHL gradient forms and (ii) validate the model with experimental data, as a proof of concept. Essentially we model the receiver cells (E. coli DH5α strain) being transformed with a plasmid containing GFP under the control of pLux promoter. Subsequently, we simulate a 2D spatio-temporal reaction-diffussion system with COMSOL Multiphysics.
Video 1: GFP expression levels over 11 hours, concentration in mol/m3. Distance between colonies: 1,5 cm.
OHHL is the only molecule in the model that is allowed to diffuse in and out cells, hence the change of concentrations over time for the other species is given by system of non-linear ordinary differential equations (ODEs). Each equation represents the rate of change of species' continuos concentration as a sum of terms representing biological processes, such as production, degradation and regulation; the later is captured with Michaelis-Menten functions. An important assumption is that specific cellular processes like transcription and translation are not modelled explicitly, but lumped together.
Our system consists of two type of genetically engineered colonies: (i) Mine Cells and (ii) Receiver Cells, that can be treated as modules that communicate only by a single molecule, OHHL. Additionally, we include the decay of OHHL while diffusing in the agar plate.
Mine cells
Two proteins of interest for our design are produced by mine cells: (i) LuxI and (ii) NagZ; both genes are constitutively expressed. Aditionally, the synthesis of OHHL is carried on by this cells; the product of luxI gene is directly involved in the synthesis of the signalling molecule, using as substrates S-adenosylmethionine (SAM) and an acylated acyl carrier protein (ACP) from the fatty acid biosynthesis pathway (Schaefer et al., 1996). We assume precursor molecules are provided by the cell in non-limiting conditions.
The system of equations for the states involved in the sender module are given below:
Agar plate
In the agar plate no reactions take place; hence only two processes of interested are considered: (i) diffusion and (ii)decay of the diffusible signal, OHHL (Eq 6).
Receiver cells
Receiver cells are responsible for processing the signal sent by the mine cells. OHHL acts as the input signal and the output is green fluorescence whose intensity correlates with the sensed OHHL concentration by receiver cells; for this purpose GFP expression is under control of pLux promoter. Once OHHL molecules reach a non-mine colony, it can bind to LuxR protein, and the complex LuxR/OHHL acts as a transcriptional activator. Therefore, this module works as a high pass amplitude filter for GFP, since it is produced when the OHHL levels are sufficiently high.
The system of equations for the states involved in the sender module are given below:
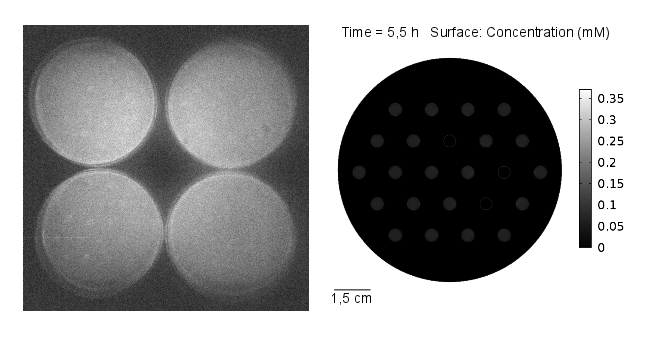
Results: simulation
The simulation was conducted for 11 hours, with 3 mine cells following the experimental setup (locations: row 2 - third colony, row 3 - fifth colony, row 4 - fourth colony). From the simulation, one can see that the predicted time scale for the expression of GFP and the experimental observations are congruent. In addition, another two observations from these results are very important:
- 1. A proper OHHL gradient can be formed to trigger expression of a protein under the control of a promoter activated by the dimer LuxR/OOHL. In this case, we can observed that the intensity of GFP correlates with the number of neighbours in the immediate vicinity.
- 2. The contribution from the basal expression of protein cannot be neglected.
Video 1: GFP expression levels over 11 hours, concentration in mol/m3. Distance between colonies: 1,5 cm.
 "
"