Team:SydneyUni Australia/Project/SinceHK
From 2013.igem.org
| (23 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
__NOTOC__ | __NOTOC__ | ||
| - | =='''Design'''== | + | =='''Post-HK Design'''== |
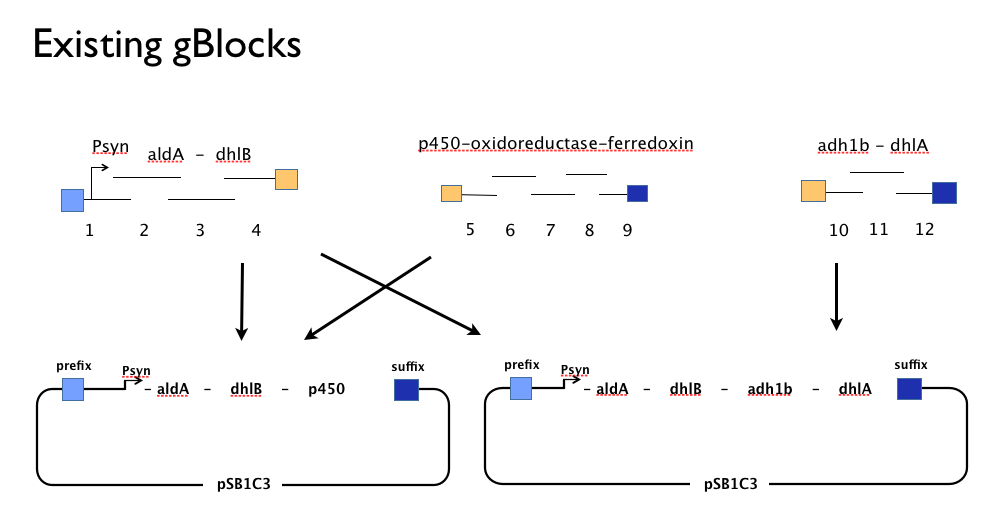
| - | Throughout the year | + | Throughout the year we have tried to assemble our DCA-degrading pathway in a single [https://2013.igem.org/Team:SydneyUni_Australia/Project/Design Gibson Assembly] reaction. An approach with which we [https://2013.igem.org/Team:SydneyUni_Australia/Project/Results had found some troubles]. We believe that the strong constitutive expression of our operon, especially some of our larger parts such as the cytochrome p450,-ferredoxin-reductase cluster may actively harm the cells or be metabolically taxing, effectively create a positive selection pressure for misassembled Gibson inserts. With this one-shot approach, we had left no flexibility in the face of failure: because we’d designed our gBlocks to assemble all at once, when this failed we couldn’t assemble the genes in our pathway from parts. |
| - | + | Having learned the virtues of modularity and flexibility the hard way, we returned from the iGEM Asia Jamboree (2013) with half-complete designs for replacement gBlocks at the start and end of each gene with the BioBrick prefix and suffix respectively. These new gBlocks, along with our old intermediate gBlocks could be gibson assembled with the Registry’s shipping vector, pSB1C3 separately. Because we still suspected that our pathway, or parts of our pathway, might be toxic in ''E.coli'' we designed an inducible promoter system consisting of the Tetracycline resistance operon promoter preceded by a constitutive tetracycline repressor protein generator. | |
| Line 22: | Line 22: | ||
This approach would allow us to: | This approach would allow us to: | ||
*use the more time-consuming but flexible of [http://parts.igem.org/Assembly:Standard_assembly conventional BioBrick assembly]. | *use the more time-consuming but flexible of [http://parts.igem.org/Assembly:Standard_assembly conventional BioBrick assembly]. | ||
| - | *identify which, if any, genes present a toxic or metabolic burden to E. coli. | + | *identify which, if any, genes present a toxic or metabolic burden to ''E. coli''. |
*characterise each gene individually to inform our [https://2013.igem.org/Team:SydneyUni_Australia/Modelling_Model model]. | *characterise each gene individually to inform our [https://2013.igem.org/Team:SydneyUni_Australia/Modelling_Model model]. | ||
*optimise the [https://2013.igem.org/Team:SydneyUni_Australia/Project/Design#order order of genes] in our pathway. | *optimise the [https://2013.igem.org/Team:SydneyUni_Australia/Project/Design#order order of genes] in our pathway. | ||
| - | *submit all of our | + | *submit all of our genes as individually characterised parts that might be used by others in any other projects or context. |
| + | =='''Post-HK Results'''== | ||
| - | |||
| - | + | ==='''Gibson Assembly'''=== | |
| - | + | We assembled the genes in our pathway individually from our new gBlocks, and screened colonies from the transformation of the Gibson Assembly reaction product. This meant we had generated clones containing the following constructs: | |
| - | * | + | |
| - | * | + | * An [http://www.ncbi.nlm.nih.gov/protein/2660722 aldehyde dehydrogenase] from ''Xanthobacter autotrophicus'' (aldA). |
| - | * | + | * The gene cluster containing [http://www.ncbi.nlm.nih.gov/protein/91700989 cytochrome p450]-[http://www.ncbi.nlm.nih.gov/protein/91700987 ferredoxin]-[http://www.ncbi.nlm.nih.gov/protein/91700988 reductase] from ''Polararomonas JS666'' (p450). |
| - | + | *Our Tet-based inducible promoter system, inspired by a tetR sequence in the Registry ([http://parts.igem.org/Part:BBa_C0040 BBa_C0040]), with our own additions including a native bi-directional Ptet promoter from [http://www.ncbi.nlm.nih.gov/pubmed/6311683 TN10]. | |
| + | *Not the human liver alcohol dehydrogenase, [http://www.ncbi.nlm.nih.gov/pubmed/16792560 adh1b2], because our supervisor forgot to order it! Oops! | ||
| + | |||
| + | The cytochrome p450-ferredoxin-reductase cluster transformants yielded comparatively fewer colonies than the other two. This may be the result of a more complex Gibson Assembly (five blocks compared to two or three). We generated more clones for screening by transforming more of the Gibson Assembly reaction product. | ||
| + | |||
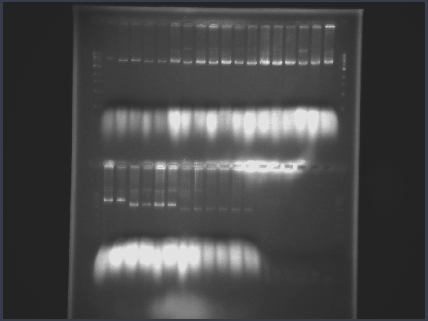
| + | We genotypically screened around 40 colonies for each construct initially by junction PCR, followed by diagnostic digests of plasmid preparations of promising colonies. | ||
| + | |||
| + | |||
| + | [[File:Whole_gel.jpg|center]] | ||
| + | |||
| + | |||
| + | <div align="justify"><div align="center"> ''We digested plasmids at RE sites contained exclusively within each gBlock. This gave us a thorough indication that all gBlocks had successfully assembled into our new parts. The entire top row and the two left-most wells of the bottom row contain 4 different p450 clones, digested with 5 different enzymes, all showing the correct band at ~5kb. The next four wells contain aldA clones, the final six contain tetR clones.''</div></div> | ||
| + | |||
| + | |||
| + | |||
| + | ==='''Cloning'''=== | ||
| + | |||
| + | We cloned our inducible promoter system in front of each transcriptional part (aldA and p450) and genotypically screened colonies by junction PCR and plasmid digest. | ||
| + | |||
| + | |||
| + | [[File:2013-10-28_TP_and_TBA__digest_looooong_stain.jpg|center]] | ||
| + | |||
| + | |||
| + | <div align="center"> ''1kb ladder, 3xTetR aldA digest with NdeI and ClaI correct construct fragments are 3424 and 2452bp''</div> | ||
| + | |||
| + | |||
| + | For the first time in our project, we used [http://parts.igem.org/Assembly:Standard_assembly conventional BioBrick assembly] to try to build our parts into a pathway, but also took advantage of the design process with gBlocks for rapid assembly. We introduced BioBrick-compatible RE sites in the beginning and end of specific parts, allowing two-way ligation into prefix and suffix regions of a BioBrick. This process makes the ligation reaction less efficient, in the sense that the host vector more frequently passes through the cloning process and is transformed amongst successful clones, but this process also saved us time. | ||
| + | |||
| + | |||
| + | |||
| + | ==='''Characterisation'''=== | ||
| + | |||
| + | |||
| + | ====''[http://aem.asm.org/content/79/7/2263.short p450]''==== | ||
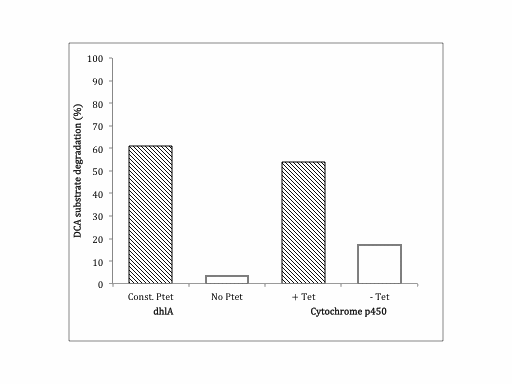
| + | This part catalyses the degradation of 1,2-dichloroethane (DCA) to chloracetaldehyde, releasing chloride ions. We performed a [https://2013.igem.org/Team:SydneyUni_Australia/Project/Protocols#chloride chloride assay] on successfully screened clones of p450. We ran our experiment in triplicate and in the presence and absence of tetracycline to test the activity of p450 on DCA with our new tet-inducible promoter system. The results below are '''VERY''' fresh and require further confirmation, but look promising. | ||
| + | |||
| + | |||
| + | [[File:Cl_Assay_Graph.png|center]] | ||
| + | |||
| + | |||
| + | We have also attempted an ethene-epoxide assay to screen for activity of the monooxygenase on non-DCA substrates, but have not found reliable characterisation yet. | ||
| + | |||
| + | |||
| + | |||
| + | ====''[http://www.sciencedirect.com/science/article/pii/S0378111997005982 aldA]''==== | ||
| + | |||
| + | This part catalyses the degradation of the chloracetaldehyde to chloroacetate, but also has activity on acetaldehyde. Both chloracetaldehyde and acetaldehyde are cytotoxic. We tried to screen for the expression of aldA with an [https://2013.igem.org/Team:SydneyUni_Australia/Project/Protocols#acetaldehyde acetaldehyde inhibition assay] by growing ''E. coli'' in the presence of acetaldehyde (at concentrations between 0-50mM), hoping to see some difference in the growth rate of our engineered cells carrying aldA. Again, the results below are fresh, but promising. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:AldA assay.gif|center]] | ||
| - | |||
{{Team:SydneyUni_Australia/Footer}} | {{Team:SydneyUni_Australia/Footer}} | ||
Latest revision as of 02:57, 29 October 2013


Post-HK Design
Throughout the year we have tried to assemble our DCA-degrading pathway in a single Gibson Assembly reaction. An approach with which we had found some troubles. We believe that the strong constitutive expression of our operon, especially some of our larger parts such as the cytochrome p450,-ferredoxin-reductase cluster may actively harm the cells or be metabolically taxing, effectively create a positive selection pressure for misassembled Gibson inserts. With this one-shot approach, we had left no flexibility in the face of failure: because we’d designed our gBlocks to assemble all at once, when this failed we couldn’t assemble the genes in our pathway from parts.
Having learned the virtues of modularity and flexibility the hard way, we returned from the iGEM Asia Jamboree (2013) with half-complete designs for replacement gBlocks at the start and end of each gene with the BioBrick prefix and suffix respectively. These new gBlocks, along with our old intermediate gBlocks could be gibson assembled with the Registry’s shipping vector, pSB1C3 separately. Because we still suspected that our pathway, or parts of our pathway, might be toxic in E.coli we designed an inducible promoter system consisting of the Tetracycline resistance operon promoter preceded by a constitutive tetracycline repressor protein generator.
This approach would allow us to:
- use the more time-consuming but flexible of [http://parts.igem.org/Assembly:Standard_assembly conventional BioBrick assembly].
- identify which, if any, genes present a toxic or metabolic burden to E. coli.
- characterise each gene individually to inform our model.
- optimise the order of genes in our pathway.
- submit all of our genes as individually characterised parts that might be used by others in any other projects or context.
Post-HK Results
Gibson Assembly
We assembled the genes in our pathway individually from our new gBlocks, and screened colonies from the transformation of the Gibson Assembly reaction product. This meant we had generated clones containing the following constructs:
- An [http://www.ncbi.nlm.nih.gov/protein/2660722 aldehyde dehydrogenase] from Xanthobacter autotrophicus (aldA).
- The gene cluster containing [http://www.ncbi.nlm.nih.gov/protein/91700989 cytochrome p450]-[http://www.ncbi.nlm.nih.gov/protein/91700987 ferredoxin]-[http://www.ncbi.nlm.nih.gov/protein/91700988 reductase] from Polararomonas JS666 (p450).
- Our Tet-based inducible promoter system, inspired by a tetR sequence in the Registry ([http://parts.igem.org/Part:BBa_C0040 BBa_C0040]), with our own additions including a native bi-directional Ptet promoter from [http://www.ncbi.nlm.nih.gov/pubmed/6311683 TN10].
- Not the human liver alcohol dehydrogenase, [http://www.ncbi.nlm.nih.gov/pubmed/16792560 adh1b2], because our supervisor forgot to order it! Oops!
The cytochrome p450-ferredoxin-reductase cluster transformants yielded comparatively fewer colonies than the other two. This may be the result of a more complex Gibson Assembly (five blocks compared to two or three). We generated more clones for screening by transforming more of the Gibson Assembly reaction product.
We genotypically screened around 40 colonies for each construct initially by junction PCR, followed by diagnostic digests of plasmid preparations of promising colonies.
Cloning
We cloned our inducible promoter system in front of each transcriptional part (aldA and p450) and genotypically screened colonies by junction PCR and plasmid digest.
For the first time in our project, we used [http://parts.igem.org/Assembly:Standard_assembly conventional BioBrick assembly] to try to build our parts into a pathway, but also took advantage of the design process with gBlocks for rapid assembly. We introduced BioBrick-compatible RE sites in the beginning and end of specific parts, allowing two-way ligation into prefix and suffix regions of a BioBrick. This process makes the ligation reaction less efficient, in the sense that the host vector more frequently passes through the cloning process and is transformed amongst successful clones, but this process also saved us time.
Characterisation
[http://aem.asm.org/content/79/7/2263.short p450]
This part catalyses the degradation of 1,2-dichloroethane (DCA) to chloracetaldehyde, releasing chloride ions. We performed a chloride assay on successfully screened clones of p450. We ran our experiment in triplicate and in the presence and absence of tetracycline to test the activity of p450 on DCA with our new tet-inducible promoter system. The results below are VERY fresh and require further confirmation, but look promising.
We have also attempted an ethene-epoxide assay to screen for activity of the monooxygenase on non-DCA substrates, but have not found reliable characterisation yet.
[http://www.sciencedirect.com/science/article/pii/S0378111997005982 aldA]
This part catalyses the degradation of the chloracetaldehyde to chloroacetate, but also has activity on acetaldehyde. Both chloracetaldehyde and acetaldehyde are cytotoxic. We tried to screen for the expression of aldA with an acetaldehyde inhibition assay by growing E. coli in the presence of acetaldehyde (at concentrations between 0-50mM), hoping to see some difference in the growth rate of our engineered cells carrying aldA. Again, the results below are fresh, but promising.
 "
"