Team:UNITN-Trento/Project/Methyl Salicylate
From 2013.igem.org
| (48 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<div class="container"> | <div class="container"> | ||
| - | + | <div class="sheet"> | |
| - | + | <span class="tn-title">Results - Methyl Salicylate </span> | |
| - | + | <p> | |
| - | + | <i>B. fruity</i> needs also a fruit ripening inhibitor. It was difficult to find a volatile molecule that could be enzymatically produced by a bacteria and also demonstrated to be an efficient ripening inhibitor. There were not many candidates to choose from and after a long search we found methyl salicylate (MeSA). Previous works suggested that MeSA inhibits the ripening of kiwifruit <span class="tn-ref"> (Aghdam M. et al., Journal of Agricultural Science. June 2011, Vol. 3, 2, pp. 149-156)</span> and tomatoes, at a concentration of 0.5 mM <span class="tn-ref">(Ding, C. and Wang, Plant Science 2003, Y. 164 pp. 589-596)</span>. | |
| - | + | </p> | |
| - | + | <p> | |
| - | + | We were happy to find out that many of the parts required to produce MeSA were already available in the registry. These parts were built by the MIT 2006 iGEM team for the project <a href="http://openwetware.org/wiki/IGEM:MIT/2006/Blurb"><i>Eau de coli</i></a>. | |
| - | + | </p> | |
| - | <img id="mesapath" style= "margin-bottom: 1em"; src="https://static.igem.org/mediawiki/2013/d/dd/Tn-2013_MeSA_path.jpg"/> | + | <img id="mesapath" style= "margin-bottom: 1em"; src="https://static.igem.org/mediawiki/2013/d/dd/Tn-2013_MeSA_path.jpg"/> |
| - | + | <span class="tn-caption" style="text-align:justify;"> <b> Figure 1: </b> this picture shows the pathway that was exploited to produce methyl salicyalte. The precursor is chorismate, a metabolic intermediate of the Shikimate pathway present in many plants and bacteria (like <i>E. coli</i> and <i>B. subtilis</i>). At first, chorismate undergoes a reaction of isomerization by the isochorismate synthase (PchA) and then the salicylate is obtained by the action of PchB, an isochorismate pyruvate lyase. Both enzymes are from the microorganism <i>Pseudomonas aeruginosa</i>. In the final part of the reaction BSMT1, a methyltransferase, transfers a methyl group from the S-adenosyl-L-methionine synthesized by the SAM synthetase. This enzyme is already present in <i>E. coli</i>. We thought that adding another copy of this gene would ultimately result in an increased MeSA production. </span> | |
| - | + | <p> | |
| - | + | We modified and improved these parts and resubmitted them to the registry. Especially, we substituted the pTet promoter controlling BSMT1 with araC-pBAD. The MIT team did not include in their MeSA generator device SAM synthetase, that we hope will boost MeSA production. We also have re-submitted in pSB1C3 each single enzyme of the pathway. | |
| - | + | </p> | |
| - | + | <img id="parts" src="https://static.igem.org/mediawiki/2013/4/41/Tn-2013_Mesa_main_parts.jpg" /> | |
| - | + | <span class="tn-subtitle">MeSA detection</span> | |
| - | + | <p> | |
| - | + | MeSA is an highly volatile liquid with a distinct minty fragrance. We exploited the physical properties of MeSA to quantify its production by gas chromatography using a Finnigan Trace GC ULTRA connected to a <b>flame ionization detector</b> (FID). This kind of instrument is able to detect ions formed during MeSA combustion in a hydrogen flame. The generation of this ions is proportional to MeSA concentration in the sample stream. A calibration curve was initially created using samples with a well known pure MeSA concentration (0 mM, 0.2 mM, 0.5 mM, 1.0 mM, 2 mM). For more details about the protocol that we used for the instrument check <a href=" https://2013.igem.org/Team:UNITN-Trento/Protocols#MeSA-detection">here</a> . | |
| - | + | </p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <div class= "tn-doublephoto-wrap"> | |
| - | + | <img class="plot" style="height: 280px; width: auto;" src="https://static.igem.org/mediawiki/2013/4/4f/Tn-2013_Taratura_MeSA.jpg"/> | |
| - | + | <img class="plot" style="height: 280px; width: auto;" src="https://static.igem.org/mediawiki/2013/7/79/Tn-2013_Pedro_GC.JPG" /> | |
| - | + | </div> | |
| - | + | <span style="text-align:justify;" class="tn-caption center"><b>Figure 3.</b> Left panel: calibration curve obtained with different concentrations of pure MeSA in ethanol. We chose to use ethanol to build up the calibration curve because at the beginning we had some issues with the solubility of the methyl salicylate. However, during our experience we performed other measurements to verify the equivalence of using ethanol and LB as a matrix for dissolving MeSA. Right panel: GC-FID in action </span> | |
| - | + | ||
| - | + | Once obtained the calibration curve, NEB10β cells transformed with <a href="http://parts.igem.org/Part:BBa_K1065102">BBa_K1065102</a> were grown both in LB and M9 medium induced with 5 mM arabinose and in some cases supplemented with salicylic acid. All the gas chromatography measurements reported here were performed in liquid phase, by injecting 1 µl of pre-filtered culture in the instrument. | |
| - | + | ||
| - | + | <img src="https://static.igem.org/mediawiki/2013/d/d4/Induced_sample_produce_MeSA.png"> | |
| - | + | <span class="tn-caption center" style="text-align:justify;"><b>Figure 4:</b> induced sample produces MeSA. A culture of NEB10β cells transformed with <a href="http://parts.igem.org/Part:BBa_K1065102">BBa_K1065102</a> was grown until O.D. 0.6 was reached. The culture was then splitted in 2 samples and one was induced with 5 mM arabinose. 2 mM salycilic acid was added to these samples. After about 4 h the samples were connected to the Gas Chromatograph. The induced sample (blue) shows the characteristic peak of methyl salicylate, as opposed to non induced cells (red).</span> | |
| - | + | ||
| + | Once we had all the chromatograms, with the software <i> Finningan Xcalibur® </i>, we were able to obtain directly the MeSA quantities from each bacterial samples. Below we report the most significant data. | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2013/3/3f/Tn-2013_Istogramma_mesa.jpg"/> | ||
| + | |||
| + | <span style="text-align:justify;" class="tn-caption center"><b>Figure 5:</b> quantification of MeSA by GC-FID. NEB10β cells transformed with <a href="http://parts.igem.org/Part:BBa_K1065102">BBa_K1065102</a> supplemented with salycilic acid produce around 0.4 mM of MeSA. Non transformed cells and non induced cells did not produce any MeSA. Cells induced with arabinose and not supplemented with salycilic acid did not show any significant MeSA concentration (data not shown).</span> | ||
| + | <span class="tn-subtitle">MeSA: 1ppm is better than zero</span> | ||
| + | <p>In addition to measurements in the liquid phase, we also tried to quantify the amount of MeSA produced by our device and able to escape in the gas phase.</p> | ||
| + | <div class="tn-doublephoto-wrap"> | ||
| + | <img class="plot"style="width:49%;"src="https://static.igem.org/mediawiki/2013/0/03/Tn-2013_BBa_K1065102_induced.jpg"/> | ||
| + | <img class="plot"style="width:49%;"src="https://static.igem.org/mediawiki/2013/a/a5/Tn-2013_BBa_K1065106_induced_%2B_SA.jpg"/> | ||
| + | |||
| + | </div> | ||
| + | <center><img class="plot"style="width:49%;border: 3px solid white; | ||
| + | box-shadow: 2px 2px 4px #323232;margin-top: -6px;"src="https://static.igem.org/mediawiki/2013/3/34/15_ppm_of_Mesa_Pure.jpg"/></center> | ||
| + | <span class="tn-caption"><b>Figure 6:</b> Quantification of MeSA by gas chromatography. (a) NEB10β cells transformed with BBa_K1065102 and induced with 5 mM arabinose, (b) NEB10β cells transformed with <a href="http://parts.igem.org/Part:BBa_K1065106">BBa_K1065106</a> and induced with 5 mM arabinose and supplemented with salycilic acid, (c) 15 ppm reference point.</span> | ||
| + | <p> | ||
| + | NEB10β cells transformed with <a href="http://parts.igem.org/Part:BBa_K1065102">BBa_K1065102</a> and <a href="http://parts.igem.org/Part:BBa_K1065106">BBa_K1065106</a> were grown in M9 medium, induced with 5 mM arabinose and in some cases supplemented with 2mM of salicylic acid. After 4 hours we performed a gas chromatography with a column optimized for the fast analysis of volatile compounds (J&W GC Column Performance Summary-Agilent Tecnologies). Peak corresponding to MeSA eluted at a retention time of 5.5 min. The quantitative analysis done by integration of the peak area showed that small amounts of MeSA are released in the gas phase under this experimental conditions: 1.3 ppm for <a href="http://parts.igem.org/Part:BBa_K1065102">BBa_1065102</a> and 0.9 ppm for <a href="http://parts.igem.org/Part:BBa_K1065106">Bba_K1065106</a> (in the presence of salicylic acid). Non induced cells did not produce any MeSA (data not shown). | ||
| + | </p><br/> | ||
| + | <span class="tn-title">Summary</span> | ||
| + | Our MeSA devices <a href="http://parts.igem.org/Part:BBa_K1065102">BBa_K1065102</a> and <a href="http://parts.igem.org/Part:BBa_K1065106">BBa_K1065106</a> were able to produce a significant concentration of MeSA only in the presence of salycilic acid. This finding was also previously observed by the MIT team in 2006 with their device (<a href="http://parts.igem.org/Part:BBa_J45700">BBa_J45700</a>). Additionally, it seems that more MeSA is present in the liquid phase than in the gas phase. <br><br> | ||
| + | After we received the DNA sequencing results of the MIT part (<a href="http://parts.igem.org/Part:BBa_J45300">BBa_J45300</a>) and of our complete device (built with MIT parts) we realized that the pLac promoter was missing the -35 box, thus generating a less strong promoter. We believe that this problem can significantly affect the correct functioning of the device. We are now in the process of improving this part by mutagenesis to rebuild a full functional pLAC promoter. | ||
| + | </div> | ||
| + | <div class="sheet-2"> | ||
| + | <a href="https://2013.igem.org/Team:UNITN-Trento/Project/Ethylene"> | ||
| + | <img class="tn-arr-prev" src="https://static.igem.org/mediawiki/2013/f/f0/Tn-2013-arr-MeSA_prev.png" /> | ||
| + | </a> | ||
| + | <a href="javascript:toTop('#tn-main-wrap-wrap');"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/6/6e/Tn-2013-arr-AAA_TOP.png" /> | ||
| + | </a> | ||
| + | <a href="https://2013.igem.org/Team:UNITN-Trento/Project/Blue%20light"> | ||
| + | <img class="tn-arr-next" src="https://static.igem.org/mediawiki/2013/2/26/Tn-2013-arr-MeSA_next.png" /> | ||
| + | </a> | ||
| + | </div> | ||
</div> | </div> | ||
<!--end content--></html>|<html>https://static.igem.org/mediawiki/2013/3/3d/Tn-2013-headerbg-Sfondowm.jpg</html>|<html>https://static.igem.org/mediawiki/2013/1/10/Tn-2013-headerbgSfondowm_or.jpg</html>}} | <!--end content--></html>|<html>https://static.igem.org/mediawiki/2013/3/3d/Tn-2013-headerbg-Sfondowm.jpg</html>|<html>https://static.igem.org/mediawiki/2013/1/10/Tn-2013-headerbgSfondowm_or.jpg</html>}} | ||
Latest revision as of 22:55, 4 October 2013
B. fruity needs also a fruit ripening inhibitor. It was difficult to find a volatile molecule that could be enzymatically produced by a bacteria and also demonstrated to be an efficient ripening inhibitor. There were not many candidates to choose from and after a long search we found methyl salicylate (MeSA). Previous works suggested that MeSA inhibits the ripening of kiwifruit (Aghdam M. et al., Journal of Agricultural Science. June 2011, Vol. 3, 2, pp. 149-156) and tomatoes, at a concentration of 0.5 mM (Ding, C. and Wang, Plant Science 2003, Y. 164 pp. 589-596).
We were happy to find out that many of the parts required to produce MeSA were already available in the registry. These parts were built by the MIT 2006 iGEM team for the project Eau de coli.
 Figure 1: this picture shows the pathway that was exploited to produce methyl salicyalte. The precursor is chorismate, a metabolic intermediate of the Shikimate pathway present in many plants and bacteria (like E. coli and B. subtilis). At first, chorismate undergoes a reaction of isomerization by the isochorismate synthase (PchA) and then the salicylate is obtained by the action of PchB, an isochorismate pyruvate lyase. Both enzymes are from the microorganism Pseudomonas aeruginosa. In the final part of the reaction BSMT1, a methyltransferase, transfers a methyl group from the S-adenosyl-L-methionine synthesized by the SAM synthetase. This enzyme is already present in E. coli. We thought that adding another copy of this gene would ultimately result in an increased MeSA production.
Figure 1: this picture shows the pathway that was exploited to produce methyl salicyalte. The precursor is chorismate, a metabolic intermediate of the Shikimate pathway present in many plants and bacteria (like E. coli and B. subtilis). At first, chorismate undergoes a reaction of isomerization by the isochorismate synthase (PchA) and then the salicylate is obtained by the action of PchB, an isochorismate pyruvate lyase. Both enzymes are from the microorganism Pseudomonas aeruginosa. In the final part of the reaction BSMT1, a methyltransferase, transfers a methyl group from the S-adenosyl-L-methionine synthesized by the SAM synthetase. This enzyme is already present in E. coli. We thought that adding another copy of this gene would ultimately result in an increased MeSA production.
We modified and improved these parts and resubmitted them to the registry. Especially, we substituted the pTet promoter controlling BSMT1 with araC-pBAD. The MIT team did not include in their MeSA generator device SAM synthetase, that we hope will boost MeSA production. We also have re-submitted in pSB1C3 each single enzyme of the pathway.
 MeSA detection
MeSA detection
MeSA is an highly volatile liquid with a distinct minty fragrance. We exploited the physical properties of MeSA to quantify its production by gas chromatography using a Finnigan Trace GC ULTRA connected to a flame ionization detector (FID). This kind of instrument is able to detect ions formed during MeSA combustion in a hydrogen flame. The generation of this ions is proportional to MeSA concentration in the sample stream. A calibration curve was initially created using samples with a well known pure MeSA concentration (0 mM, 0.2 mM, 0.5 mM, 1.0 mM, 2 mM). For more details about the protocol that we used for the instrument check here .


 Figure 4: induced sample produces MeSA. A culture of NEB10β cells transformed with BBa_K1065102 was grown until O.D. 0.6 was reached. The culture was then splitted in 2 samples and one was induced with 5 mM arabinose. 2 mM salycilic acid was added to these samples. After about 4 h the samples were connected to the Gas Chromatograph. The induced sample (blue) shows the characteristic peak of methyl salicylate, as opposed to non induced cells (red).
Once we had all the chromatograms, with the software Finningan Xcalibur® , we were able to obtain directly the MeSA quantities from each bacterial samples. Below we report the most significant data.
Figure 4: induced sample produces MeSA. A culture of NEB10β cells transformed with BBa_K1065102 was grown until O.D. 0.6 was reached. The culture was then splitted in 2 samples and one was induced with 5 mM arabinose. 2 mM salycilic acid was added to these samples. After about 4 h the samples were connected to the Gas Chromatograph. The induced sample (blue) shows the characteristic peak of methyl salicylate, as opposed to non induced cells (red).
Once we had all the chromatograms, with the software Finningan Xcalibur® , we were able to obtain directly the MeSA quantities from each bacterial samples. Below we report the most significant data.
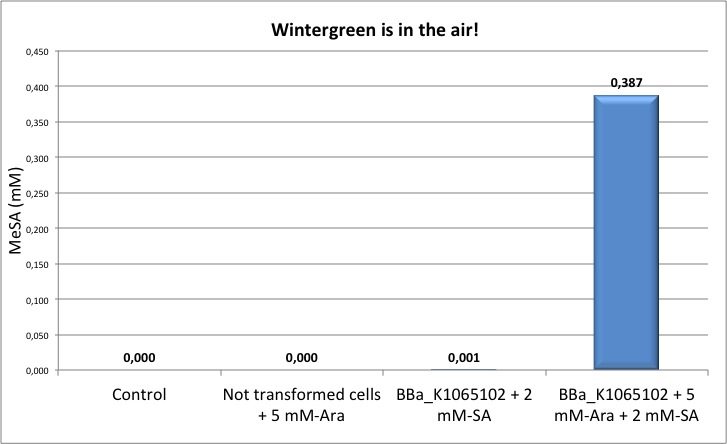 Figure 5: quantification of MeSA by GC-FID. NEB10β cells transformed with BBa_K1065102 supplemented with salycilic acid produce around 0.4 mM of MeSA. Non transformed cells and non induced cells did not produce any MeSA. Cells induced with arabinose and not supplemented with salycilic acid did not show any significant MeSA concentration (data not shown).
MeSA: 1ppm is better than zero
Figure 5: quantification of MeSA by GC-FID. NEB10β cells transformed with BBa_K1065102 supplemented with salycilic acid produce around 0.4 mM of MeSA. Non transformed cells and non induced cells did not produce any MeSA. Cells induced with arabinose and not supplemented with salycilic acid did not show any significant MeSA concentration (data not shown).
MeSA: 1ppm is better than zero
In addition to measurements in the liquid phase, we also tried to quantify the amount of MeSA produced by our device and able to escape in the gas phase.



NEB10β cells transformed with BBa_K1065102 and BBa_K1065106 were grown in M9 medium, induced with 5 mM arabinose and in some cases supplemented with 2mM of salicylic acid. After 4 hours we performed a gas chromatography with a column optimized for the fast analysis of volatile compounds (J&W GC Column Performance Summary-Agilent Tecnologies). Peak corresponding to MeSA eluted at a retention time of 5.5 min. The quantitative analysis done by integration of the peak area showed that small amounts of MeSA are released in the gas phase under this experimental conditions: 1.3 ppm for BBa_1065102 and 0.9 ppm for Bba_K1065106 (in the presence of salicylic acid). Non induced cells did not produce any MeSA (data not shown).
Summary Our MeSA devices BBa_K1065102 and BBa_K1065106 were able to produce a significant concentration of MeSA only in the presence of salycilic acid. This finding was also previously observed by the MIT team in 2006 with their device (BBa_J45700). Additionally, it seems that more MeSA is present in the liquid phase than in the gas phase.
After we received the DNA sequencing results of the MIT part (BBa_J45300) and of our complete device (built with MIT parts) we realized that the pLac promoter was missing the -35 box, thus generating a less strong promoter. We believe that this problem can significantly affect the correct functioning of the device. We are now in the process of improving this part by mutagenesis to rebuild a full functional pLAC promoter.
 "
"
















