IDEA!
Place sgRNAs under control of different light sensors. One light one sgRNA.
Our system operates like this:
1. light sensors regulates sgRNAs;
2. then sgRNAs regulate their corresponding genes, which is the final targets in the pathway to be optimized.
Therefore, we are able to:
In the past, researchers seldom accomplishes the regulation of genomic genes since it is so difficult to change endogenous promoters to be regulated by outer signals.
- Simultaneously controlling several genes.
Each gene is regulated by a different light, respectively.
- Offer a easy-to-use platform.
Users of our Metabolic Gear Box can optimize their own desired pathway after making some minor changes to a vector of sgRNAs.
Testing CRISPRi
To test CRISPRi, we have constructed a constitutive dCas9 on pRSFDuet, a constitutive sgRNA targeting mRFP (gR4mRFP) on pCDFDuet and a constitutive mRFP on pETDuet.
Then we set up a strictly controlled experiment as follows:
- Case: constitutive dCas9 on pRSFDuet + a constitutive gR4mRFP on pCDFDuet + constitutive mRFP on pETDuet
- Controll: constitutive dCas9 on pRSFDuet + an empty pCDFDuet + constitutive mRFP on pETDuet
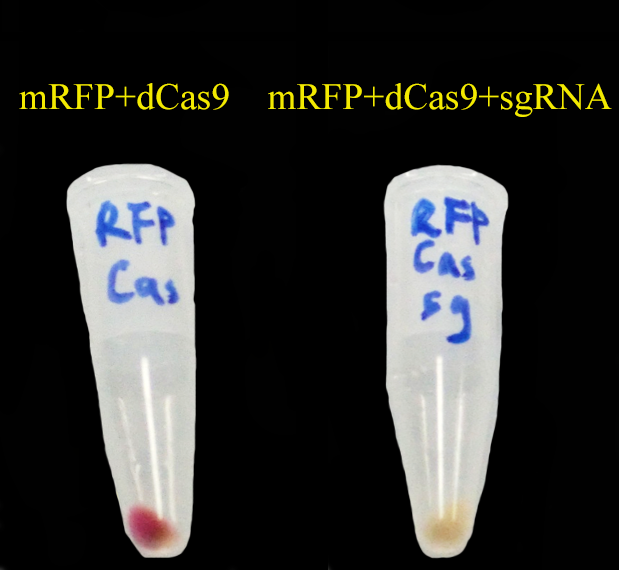
As expected, mRFP repressed by CRISPRi (dCas9 and gR4mRFP) is not as red as the control.
The sequence of gR4mRPF is given in our part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1026003 BBa_K1026003]. This sequence comes from the following literature: QI, LEI S., LARSON, MATTHEW H., GILBERT, LUKE A., DOUDNA, JENNIFER A., WEISSMAN, JONATHAN S., ARKIN, ADAM P. & LIM, WENDELL A. 2013. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell, 152, 1173-1183.
And CAUTION that:
- There shall not be extra nucleotides between promoter and sgRNA itself.
gRNA shall be exactly transcribed out. Any extra nucleotides would potentially jeopardize the normal function of the gRNA. Therefore, BioBrick scars are not allowed here. Instead of cut-and-ligation, inverse PCR or In-Fusion is preferred for the assembly task.
Our part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1026003 BBa_K1026003] can be an example of no-scar assembly.
- A reliable terminator is applied here to ensure the termination of sgRNA.
Testing the Interface of CRISPRi and Light Sensors
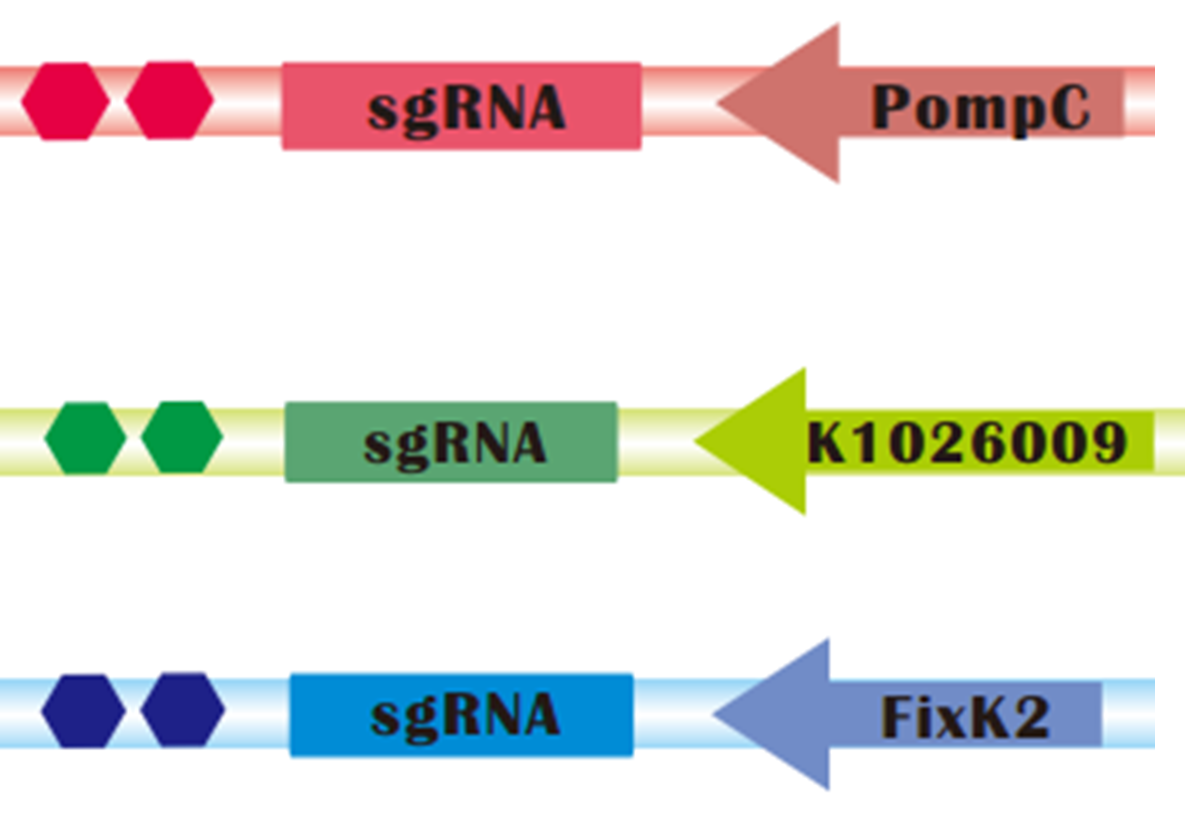 sgRNAs regulated by light sensors. Up: Red light controlled sgRNA; Medium: Green light controlled sgRNA; Down: Blue light controlled sgRNA
To assure that CRISPRi can be successfully combined with Light Sensors, we constructed sgRNAs targeting mRFP that are regulated by red, green and blue light sensors, respectively:
- POmpC is a promoter negatively regulated by Red Light.
- K1026009, one of our parts, is a artificial promoter negatively regulated by green light.
- FixK2 is a promoter negatively regulated by Blue Light.
And next, it is show time for our Metabolic Gear Box!
 A Metabolic Gear Box in work! With bacteria culture! And in a thermostat incubator!  A Metabolic Gear Box Receiving Feed-forward Commands. Researchers will now send commands of desired expression level to the computer. Then these commands will be transferred to the Box
Since it is difficult for RFP reporter to accurately produce qualitative results, we continue to the next phase of our project -- luciferase reporter assay.
Gathering Quantitative Data -- sgRNA targeting luciferase
In order to know whether our system provides a wide enough range of continuous adjustment, we constructed sgRNAs targeting luciferase (gR4luciferase) gene. And again, these gR4luciferase are under controll of red, green and blue light, respectively.
Designing a sgRNA targeting luciferase
Since we do not know the sequence of a working sgRNA that targets luciferase, so we designed two gR4mRFP that complements the coding sequence of luciferase gene ourselves according to those design criteria: one is closer to the start codon (the proximal one), whereas the other is relatively further (the distal one).
However it is necessary for us to test these sgRNA. So we again inserted these two sgRNAs into a constitutive operon. After conducting a luciferase reporter assay with a Beyotime kit, it comes out that the repression effect of these two sgRNAs are 80% and 60%, respectively.
Therefore, we choose the proximal one for later use.
Determining the Working Curve of our Metabolic Gear Box
Again, it is time for our Metabolic Gear Box.
|