Team:Newcastle/Project
From 2013.igem.org
YDemyanenko (Talk | contribs) |
|||
| Line 43: | Line 43: | ||
One crucially important feature of our naked bacteria is that they are osmotically sensitive, meaning that they will lyse if they escape into the environment. This means that they can be used in non-contained environments (e.g. agriculture). | One crucially important feature of our naked bacteria is that they are osmotically sensitive, meaning that they will lyse if they escape into the environment. This means that they can be used in non-contained environments (e.g. agriculture). | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
For our research we used pMutin4 plasmids. However before deciding to use this particular plamid we have tested a plasmid designed by the Groningen 2012 iGEM team. We were unable to use the plasmid and we have thus managed to [http://parts.igem.org/Part:BBa_K818000:Experience characterise it] and speculate why this part is non-functional. | For our research we used pMutin4 plasmids. However before deciding to use this particular plamid we have tested a plasmid designed by the Groningen 2012 iGEM team. We were unable to use the plasmid and we have thus managed to [http://parts.igem.org/Part:BBa_K818000:Experience characterise it] and speculate why this part is non-functional. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Modelling== | ==Modelling== | ||
| - | + | This step is an essential part of a successful synthetic biology project. Although it requires a lot of time and effort, and therefore is often neglected. We believe that in the long run modelling can save a lot of time, effort and resources to those who take their time in the beginning, simulating all the possible outcomes of the system and refining it at an early stage, before any ''in vitro'' and ''in vivo'' experiments have been planned and conducted. Another positive side to modelling prior to the "wet lab" sessions is the fact that a model behaves according to the known facts and principles, and if in real life the outcome drastically differs from the simulation, there's a good chance of finding out what may be causing the difference through adjusting the model and repeating the experiments. | |
For every research theme we have constructed a model to help us understand the systems we engineered. Click on the links to view each model or visit our [https://2013.igem.org/Team:Newcastle/Modelling modelling page]: | For every research theme we have constructed a model to help us understand the systems we engineered. Click on the links to view each model or visit our [https://2013.igem.org/Team:Newcastle/Modelling modelling page]: | ||
Revision as of 20:16, 4 October 2013

Contents |
Project Overview
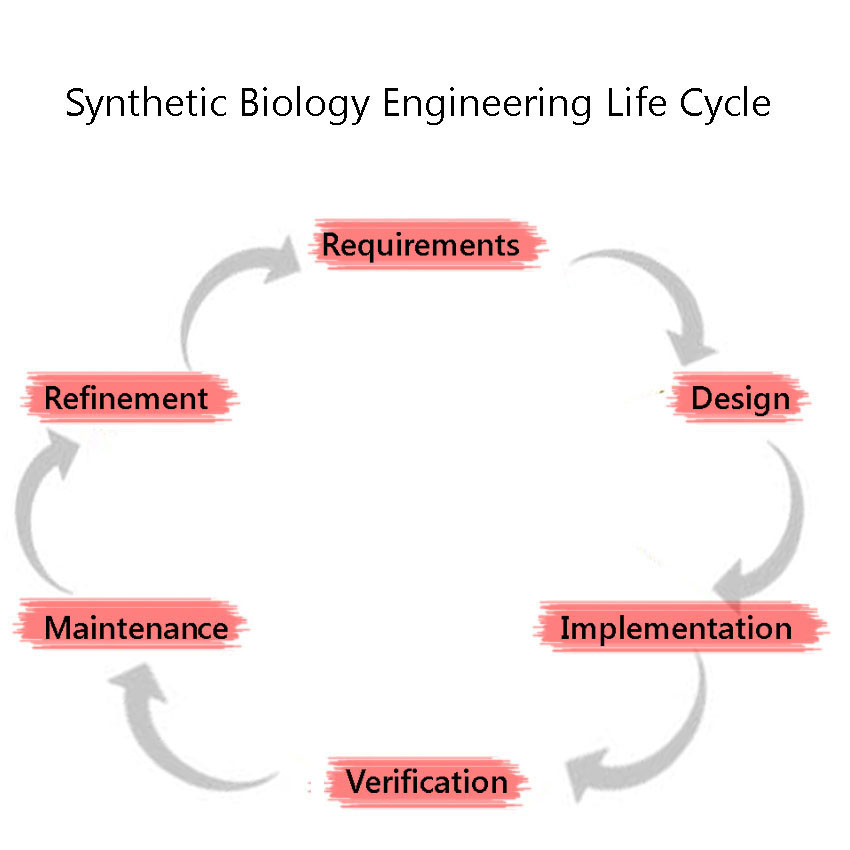
Synthetic Biology is a discipline which heavily employs engineering principles. One of these principles is the Engineering Lifecycle, a framework in which the project is split into clearly defined sections, based on the development of the project. These are, in sequential order: 1) Requirements 2) Design (including Modelling) 3) Implementation 4) Verification 5) Maintenance 6)Refinement and the cycle is iterated through again, as we attempt to improve the system further. We adhered to this cycle throughout our project including the development and characterisation of our BioBricks.
A Foundational Advance
While researching Synthetic Biology we found that a cell wall, one of the standard components of the bacterial cell, often causes difficulties in many techniques. These included transformation efficiency, secretion of recombinant proteins, adaption to the environment etc. So we thought to ourselves: is there any way to remove the cell wall and still have a viable cell?
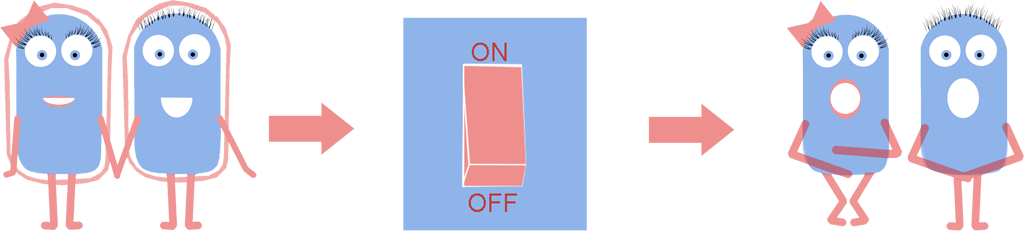
As a result we have developed a new chassis with the potential to revolutionise how Synthetic Biology is performed. The main BioBrick(BBa_K1185000) that we have introduced enables the switching on and off of the bacterial cell wall in the model Gram positive bacteria Bacillus subtilis, at the demand of the synthetic biologist, while still allowing cells to grow and divide. Employing bacterial cells without a cell wall can both enable the synthetic biologist to explore new applications and research areas, and also build-upon and improve areas that are already being explored in Synthetic Biology. Rather than the application-oriented nature of many iGEM projects, we think that the use of cell wall-less bacteria as a novel chassis in Synthetic Biology, as we propose, can benefit across the whole subject area, and furthermore be utilised as a tool to allow for even greater feats to be achieved by future iGEM teams.
Bacteria which have lost their cell wall yet are still able to grow and divide are called L-forms, or as we prefer to call them, naked bacteria. If you would like to learn more about L-forms, please take a look at our L-form page or click [http://www.youtube.com/watch?v=b0Kk6bKKOQ0 here] to watch a short video summarising our project.
Once we had created L-forms with our Switch BioBrick, we were unable to ignore the new opportunities that L-forms bring to Synthetic Biology:
Genome Shuffling
We investigated using genome shuffling and L-forms to artificially evolve B. subtilis. This technique increases the rate of evolution, allowing the improvement of any biological system or phenotype in a feasible timeframe. Over the past eight years iGEM teams have dreamt up innovative ways of harnessing Synthetic Biology. However this relatively new field faces challenges such as producing high quality yield of the desired product. Using L-forms allows the use of genome shuffling to solve this problem. If harnessed this could improve the efficiency of hundreds of iGEM projects as well as cell factories across the field of synthetic biology.
Introduction and Detection of Naked Bacteria in Plants
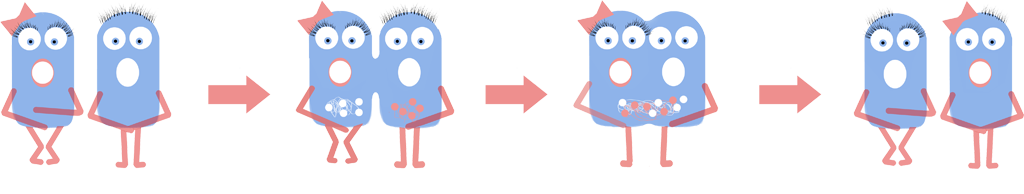
L-forms have been shown to form symbiosis in plants. Plants with naked bacteria show increased resistance to fungus and they could be used to deliver useful compounds to the plant. This could give better crop yields, more nutritious harvests and reduce the need for spraying of fertiliser, pesticides or other compounds.
Shape Shifting
The loss of the cell wall leaves L-forms protected by only a cell membrane. The plasma membrane of L-forms is quite fluid. The advantage of this is that these cells would be able to adapt to shapes of various cracks and cavities, or will be able to "squeeze through" tiny channels and deliver cargo to hard-to reach targets.
This isn’t a finite list of what can be done with naked bacteria, there’s loads more! L-forms are currently used to discover novel antibiotics which don’t act on the cell wall. L-forms can also teach us a great deal about how bacterial life has evolved, through acting as a model for a cell wall-less bacterial progenitor, and through being able to test the ease of induction of endosymbiosis in cell wall-less organisms (Mercier et al. 2013).
L-form bacteria can be used in any process which protoplasts (bacteria which have been chemically induced to lose their cell wall but which are not alive) are currently used for such as transforming bacteria which are difficult to transform (Chang and Cohen 1979).
One crucially important feature of our naked bacteria is that they are osmotically sensitive, meaning that they will lyse if they escape into the environment. This means that they can be used in non-contained environments (e.g. agriculture).
For our research we used pMutin4 plasmids. However before deciding to use this particular plamid we have tested a plasmid designed by the Groningen 2012 iGEM team. We were unable to use the plasmid and we have thus managed to [http://parts.igem.org/Part:BBa_K818000:Experience characterise it] and speculate why this part is non-functional.
Modelling
This step is an essential part of a successful synthetic biology project. Although it requires a lot of time and effort, and therefore is often neglected. We believe that in the long run modelling can save a lot of time, effort and resources to those who take their time in the beginning, simulating all the possible outcomes of the system and refining it at an early stage, before any in vitro and in vivo experiments have been planned and conducted. Another positive side to modelling prior to the "wet lab" sessions is the fact that a model behaves according to the known facts and principles, and if in real life the outcome drastically differs from the simulation, there's a good chance of finding out what may be causing the difference through adjusting the model and repeating the experiments.
For every research theme we have constructed a model to help us understand the systems we engineered. Click on the links to view each model or visit our modelling page:
Implementation
Click here to find a chronological description of things we have done and methods we've used to create and characterise our BioBricks.
Verification
By the end of our project we have:
- created and characterised the BioBrick which allows the switching on and off of the cell wall of ‘’B.subtilis”.
- Shown that the naked bacteria that we created using our switch BioBrick also form these associations.
All you need to start using an L-form chassis is a culture of Bacillus subtilis, our L-form switch BioBrick and a set of instructions from us.
Maintenance
We realised that our project has some ethical implications and therefore, have dedicated some of our time and resources to research and explore this topic.
References
[http://www.ncbi.nlm.nih.gov/pubmed/11849491 Walker R, Ferguson CMJ, Booth NA and Allan EJ (2002) The symbiosis of Bacillus subtilis L-forms with Chinese cabbage seedlings inhibits conidial germination of ‘Botrytis cinerea. Letters in Applied Microbiology, 34, 42-45.]
[http://www.ncbi.nlm.nih.gov/pubmed/107388 Chang S and Cohen SN (1979)High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Molecular Genetics & Genomics, 168, 111–115.]
[http://www.cell.com/abstract/S0092-8674(13)00135-9 Mercier R, Kawai Y and Errington J. (2013) Excess Membrane Synthesis Drives a Primitive Mode of Cell Proliferation, Cell, 152, 997–1007.]
 "
"