Team:NCTU Formosa/project
From 2013.igem.org
A multiple regulated-system was built using three different regulation mechanisms including red light, temperature, and sRNA. In other words, it is multitasking genetic engineered machine that can express a variable genes depending on the different command given.
Contents |
Introduction
It is not a dream any more that bacteria engineered machines give us the opportunity to fulfill our mission and so, we thought that one day, we could demand bacteria to work specifically along with different transition of environmental conditions. Our basic concept is ,“Each work works under a specific condition”. We use three regulating factors, light, temperature and sRNA to design a model that is cross-regulated and is able to work under different given environmental conditions. Through pairing specific conditions, we will only have to change different reacting conditions for turning on or shutting down reactions, furthermore, we use the sRNA inhibiting system to turn off unnecessary reactions. With noninvasive regulators and an easy way to adjust, such system controlled by multiple regulators is believed to be used in different applications that require specific control.
Advantages
Direct sensor
The light regulated-system is different from previous promoters, because it can detect light directly, in other words, it acts as a direct sensor.
Triggers under certain temperature
The temperature system acts as a switch which triggers under certain temperature.
Specifically binding
The function of sRNA recognizes specific target sequence and reduces mRNA transcription level. We try to use sRNA which specifically regulates our biobrick.
Noninvasive
Traditional methods we regulate gene expression, we usually use invasive chemical substance, but in our project, we want to construct a system with noninvasive by light, temperature and sRNA.
Adjustible
To construct an adjustable system, we designed these biobricks to help people replace the downstream genes they want.
sRNA
Principle and Why we use it
(undetermined)
Design
(undetermined)
Small RNAs (sRNAs) have become increasingly significant in playing the role of bacterial gene regulation. Most sRNAs interact with the targeted mRNAs by imperfect base pairing, reducing the translation efficiency and recruiting chaperones such as Hfq for translation termination. In other words, sRNAs regulate gene expression by forestalling translation. It is an effective regulated-system that functions under RNA level. Since this regulated-system has already been employed in vivo, it is necessarily to design artificial sRNA that targets specifically to the desired genes. This way, we can prevent the sRNA from effecting undesired genes.
The sRNA employed in this project was picked from a library of artificial sRNA that was constructed by fusing a randomized antisense domain of Spot42, the scaffold that is known to recruit the RNA chaperons. The sRNA we picked contains a consensus sequence, 5’-CCCUC-3’, that can base pair with the SD sequence due to complementarity. This sRNA, as expected, effectively regulates gene expression by reducing translation efficiency and recruiting Hfq. In addition, this sRNA shows high specificity against its targeted gene, ompF, as it doesn’t hold significant activity against other genes from E. Coli genome (Sharma and others, 2011).
The sRNA picked is competent, but we hoped that it can target any desired gene. With that said, we designed a RBS by employing the sRNA targeting region from ompF and making the AUG codon sufficiently apart from the SD sequence for ribosome binding. By adding this RBS to the upstream of any desired gene, the gene can be regulated by sRNA.
37 degree Celsius RBS
Principle and Why we use it
(unfinished)
Unlike normal RBS, 37°C RBS has a unique hairpin secondary structure that encloses the Shine Dalgarno site (SD site), preventing ribosome starting translation. Such structure is favored as it lowers the free energy of the RBS. And since the structure is sustained by base pairing, heat can be employed to break the hydrogen bonds. With the bonds broken, the hairpin would be unfolded, causing the SD site to be exposed and available for ribosome binding. This means that by raising temperature to a certain threshold, 37°C RBS can function as a normal RBS. The threshold of 37°C RBS employed, K115002, is 37°C , implying that K115002 would remain as hairpin structure under 37°C and only functions normally above or at 37°C . This way, the expression of the genes downstream of 37°C RBS can be regulated by temperature.
Design
(undetermined)
Light control
Mechnism
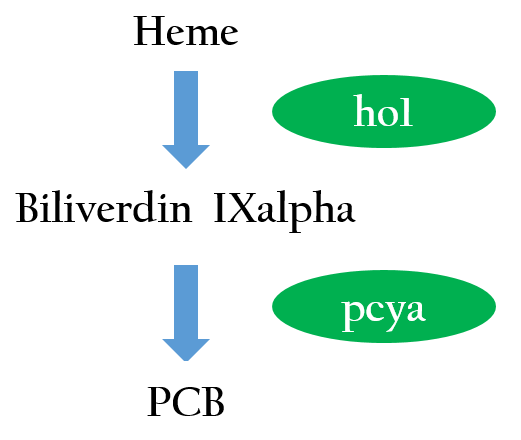
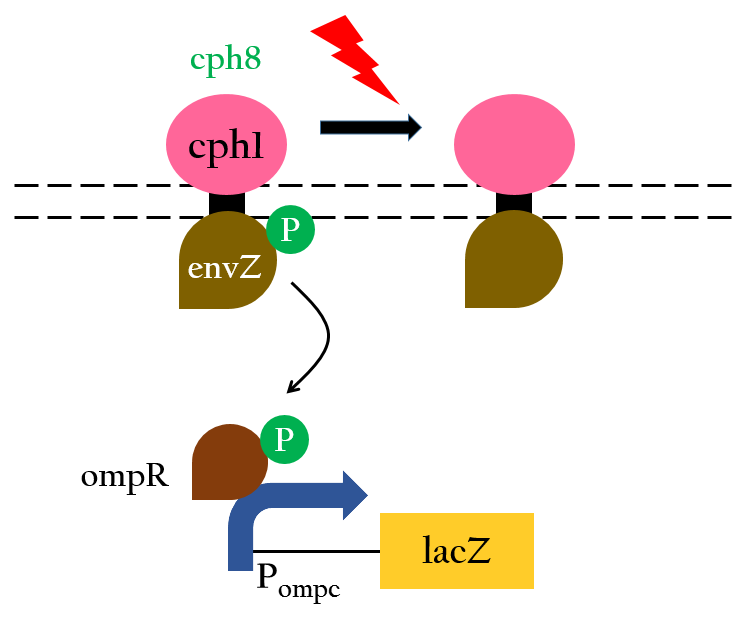
To positively control our biobricks using red light, we had to first synthesize a chlorophyll a/b protein called PCB that acts as an antenna to capture and deliver the energy of light. PCB can be converted from heme in vivo by two enzymes known as pcyA and ho1. By binding PCB with a two component protein containing histidine kinase, cph8, a red light photoreceptor is produced. It phosphorylates ompR, regulator of Pompc, to activate the promoter in absence of red light.
Pred promoter
Why we use it
(undetermined)
Design
(unfinished)
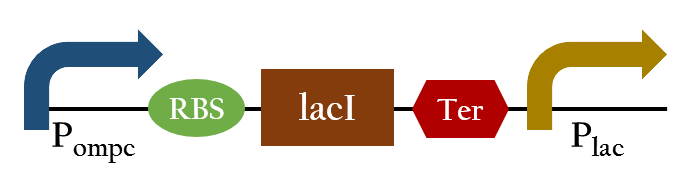
Upon exposure of red light, the phosphorylation activity is hindered, and therefore, the promoter remains repressed and the downstream genes cannot be translated. Such negative control, however, is more complicated and less straightforward than a simple positive control. With that said, we converted negative control of Pompc into positive control by building a biobrick called Pred.
In the presence of red light, Pompc is shut down and lacI cannot be expressed to repress Plac. As a result, Plac is activated and all the downstream genes can be expressed. In the absence of red light, whatsoever, Pompc is activated as phosphorylation is triggered and lacI is expressed. Consequently, lacI respressed Plac and the genes downstream cannot be expressed. By employing Pred, we can achieve positive control using red light,
Mechanism of light induced-device
Pathway Regulation
This is the pathway we designed by using the three different regulating factors. We tend to set up four conditions for the E.coli to work by paring the light and the temperature controls. We use four different fluorescent proteins as reporter genes to test that whether the switch of our pathway is successful.
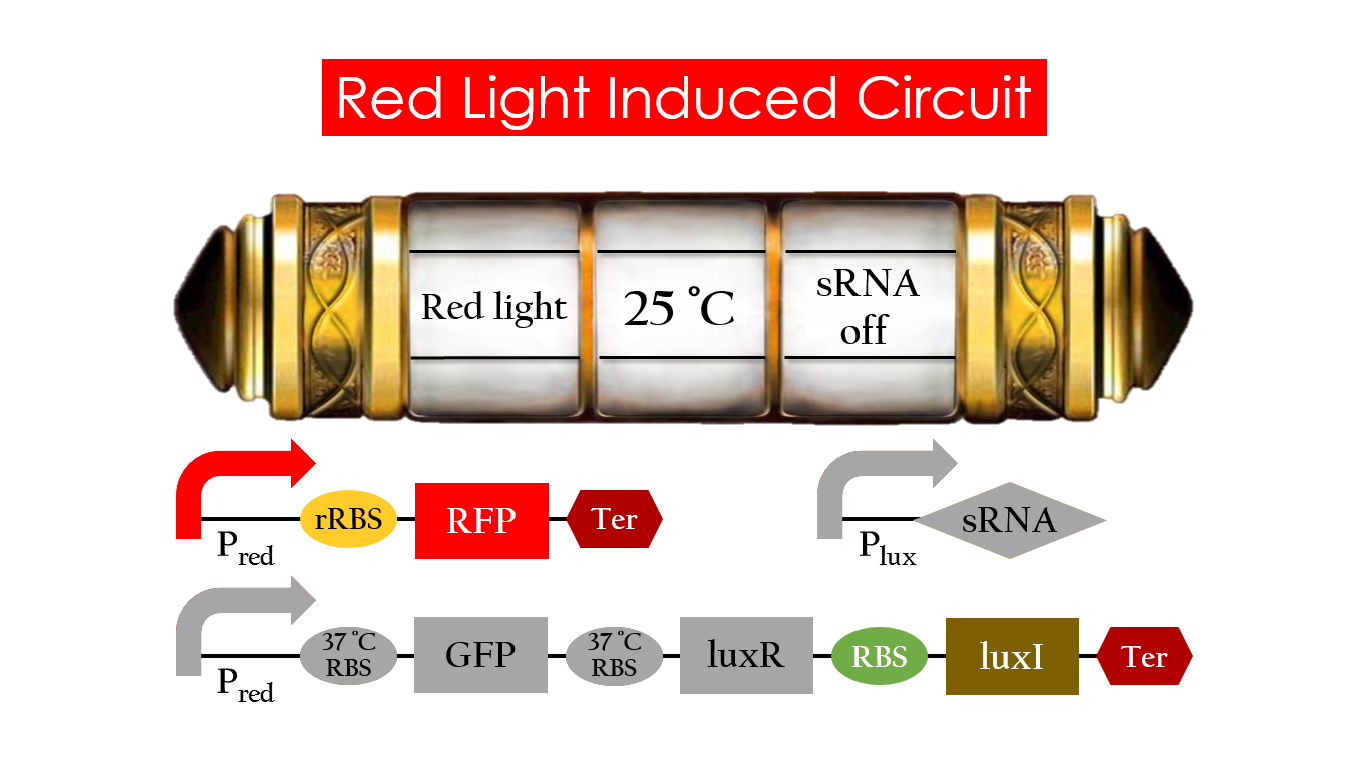
Red Light Induced Circuit
At 30°C , Pred is activated by red light and translation proceeds. However, ribosomes can only bind to the normal RBS, as 37°C RBS forms a hairpin structure, forestalling ribosomes from binding. The only gene downstream of the normal RBS is RFP, so only RFP would be expressed.
On the other hand, 37°C RBS unfolds at 37°C , resulting in the expression of both LuxR and GFP. LuxR would binds with AHL to form a complex that activates Plux. The activation of Plux produces sRNA that binds to the normal RBS, blocking it from ribosomes. As a result, RFP would not be expressed and only GFP remains at 37°C .
Without red light, Pred would not be activated, and therefore, this system is completely shut down in the dark.
Dark Response Circuit
Pred activates in the presence of red light, producing tetR that represses Ptet. In other words, the dark induced part is inactive in the exposure of red light and active without red light. At 30°C in the dark, only normal RBS functions to express YFP while 37°C RBS remains as a hairpin structure. At 37°C, however, 37°C RBS unfolds to express LuxR and BFP. Just like the mechanism employed in the light induced part, LuxR forms a complex with AHL to activate Plux that produces the sRNA to block normal RBS. This way, the YFP downstream of YFP cannot be expressed and BFP is expressed at 37°C
Future Work
The multiple regulated-system we have created in this project consisted of two different parts: light induced part and dark induced part. Each part regulates two different genes, resulting in a total of four genes that can be regulated. By adding new parts to the system, it is plausible that the system can regulate more genes. We tend to do that by employing more light sensing promoters. Each light sensor we add, we would be able to regulate two more different genes by employing regulation mechanism of 37°C RBS and sRNA.
On the other hand, we tend to optimize our system to achieve subtle control of the four gene expressions. We might be able to do this by taking the values between the limits we have set. Instead of 30°C and 37°C , we tend to take the values between so we can create a lot more conditions under which different level of expression can be achieved. We can also vary the intensity of red light to control the level of expression. The ultimate goal is to precisely control the level of expression of each gene.
Application
(undetermined)
Reference
- Guillermo Rodrigo, Thomas E. Landrain, and Alfonso Jaramillo.(2012).De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells. PNAS, 109:15271–15276
- Jennifer B. Rice, Divya Balasubramanian, and Carin K. Vanderpool.(2012).Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA.PNAS,109: E2691–E2698 DOI 10.1073/pnas.1207927109
- JoÈrg Vogel, Verena Bartels, Thean Hock Tang, Gennady Churakov, Jacoba G. Slagter-JaÈger, Alexander HuÈttenhofer, and E. Gerhart H. Wagner.(2003). RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Research.31:6435-6443
- Jamie Richards,and Joel G. Belasco.(2008). A New Window onto Translational Repression by Bacterial sRNAs. Molecular Cell 32
- Johannes H. Urban and Jörg Vogel.(2007). Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Research, 35:1018-1037 DOI:10.1093/nar/gkl1040
- Kathrin S Fröhlich and Jörg Vogel.(2009). Activation of gene expression by small RNA. Current Opinion in Microbiology, 12:674–682
- Gisela Storz, Jörg Vogel, and Karen M. Wassarman.(2011). Regulation by Small RNAs in Bacteria:Expanding Frontiers. Molecular Cell 43
- Karen M. Wassarman.(2002). Small RNAs in Bacteria: Diverse Regulators of Gene Expression in Response to Environmental Changes. Cell,109: 141–144
- Susan Gottesman.(2004). THE SMALL RNA REGULATORS OF ESCHERICHIA COLI: Roles and Mechanisms. Annu. Rev. Microbiol, 58:303–28 DOI:10.1146/annurev.micro.58.030603.123841
- Karen M. Wassarman and Ruth M. Saecker.(2006). Synthesis-Mediated Release of a Small RNA Inhibitor of RNA Polymerase. Science 314, 1601 DOI:10.1126/science.1134830
- Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park& Sang Yup Lee.(2013).Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nature Biotechnology,31:170-176 DOI:10.1038/nbt.2461
- Erel Levine, Zhongge Zhang, Thomas Kuhlman, Terence Hwa.(2007).Quantitative Characteristics of Gene Regulation by Small RNA. PLoS Biology, 5:e229
- Shuai Man, Rubin Cheng, Cuicui Miao, Qianhong Gong, Yuchao Gu, Xinzhi Lu, Feng Han and Wengong Yu.(2011). Artificial trans-encoded small non-coding RNAs specifically silence the selected gene expression in bacteria. Nucleic Acids Research,39:e50 DOI:10.1093/nar/gkr034
- Hongmarn Park, Geunu Bak, Sun Chang Kim, and Younghoon Lee.(2013). Exploring sRNA-mediated gene silencing mechanisms using artificial small RNAs derived from a natural RNA scaffold in Escherichia coli. Nucleic Acids Research,41: 3787–3804 DOI:10.1093/nar/gkt061
- Vandana Sharma, Asami Yamamura, and Yohei Yokobayashi.(2012).Engineering Artificial Small RNAs for Conditional Gene Silencing in Escherichia coli. ACS Synth. Biol.,1:6-13
 "
"