Team:NYMU-Taipei/Modeling/MainParts
From 2013.igem.org


Contents |
main part
Function of the parts:
circuit regulators:
• LacI regulated promoter(pLac) : when LacI exists, it will bind to LacI regulated promoter(pLac) and represses the promoter.
• pLux/cI hybrid promoter(plux/cI) : when luxR/AHL exists, it will open pLux/cI hybrid promoter(plux/cI), while cI will repress the promoter. What’s more, plux/cI is cI-dominant, which means when cI exists, the hybrid promoter will be repressed whether luxR/AHL exists or not.
Circuit regulation:
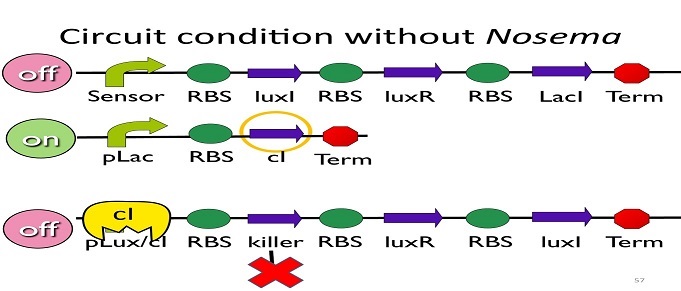
• Circuit condition without Nosema:
Without Nosema Ceranae, the first circuit will not open, and thus, no LacI will be produced. Since there is no LacI binding to pLac, pLac will not be repressed, which means cI will be produced. After that, cI will bind to pLux/cI hybrid promoter, leading the third circuit to be off.
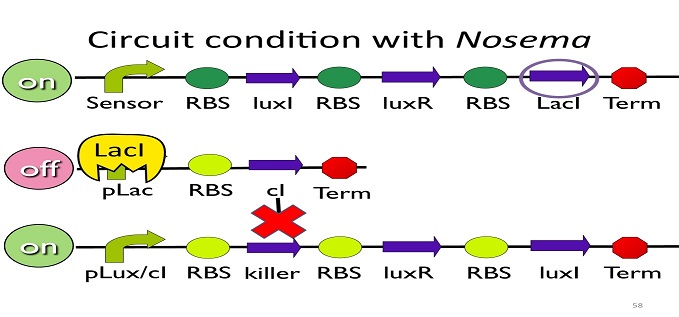
• Circuit condition with Nosema:
When Nosema Ceranae infects the bee, bee’s immune system will be activated, leading to the concentration of ROS (reactive oxygen species) to be high. In response, Bee. coli will enhance the production of oxyR, which forms a complex with ROS and binds to transcription binding site ahead of trxC (an oxyR-activated promoter; namely, the sensor), leading to the first circuit to be on.
Since the first circuit is on, the downstream LacI gene will be produced and bind to pLac, leading to the second circuit to be off.
After the second circuit is closed, no cI is produced, leading to the third circuit to be on (no repressor at all). And then, kill protein will be produced and kill Nosema Ceranae.
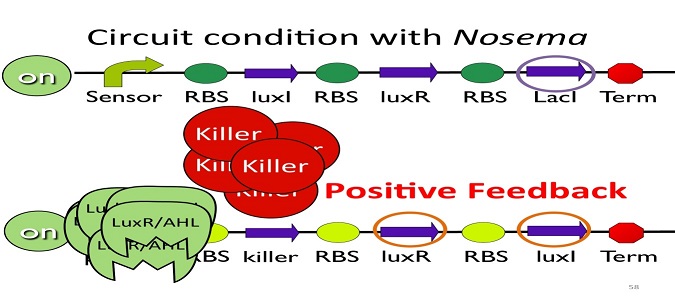
•Positive feedback:
Besides the LuxI and LuxR(which then form a complex called luxR/AHL) produced by first circuit, the third circuit will also produce LuxI and LuxR, which acts as a positive feedback and strongly enhances the production of killer protein.
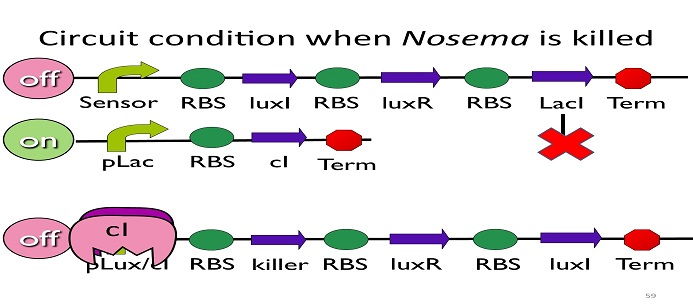
•Circuit condition when Nosema is killed:
After Nosema is killed, the sensor will be off and no lacI will be produced.
Without LacI, the pLac will not be repressed, and the second circuit will be on, leading to the production of cI. After cI binds to pLux/cI hybrid promoter, the third circuit to be off. After that, the overall circuit will switch to the beginning stage when there is no Nosema.
Aims
#To know how much time it needs from sensing to killing the Nosema after infection #To know whether the pathway is effective or not #To know the range of kill protein concentration (from the minimal concentration which Is effective to kill Nosema to the maximal concentration which will not do harm to the bees)
It is assumed that AHL is abundant and thus the formation of AHL2LuxR complex is determined only by the concentration of LuxR; CI’s mechanism of binding to the promoter is assumed to act as hill effect form; kill protein will sustain a period of time before it is degraded naturally without any other types of decomposition.
Equations
Equation1:
KdLacI = dissociation constant of CI
nLacI = Hill coefficient of LacI
PoPSpLac = promoter strength of pLac
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNACI production rate and when it can reach the level to translate the desired concentration.
Equation2:
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPStrxC = promoter strength of trxC
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how trxC promoter strength (in PoPS) influences the production of lacI.
Equation3:
PoPSconstitutive= promoter strength of constitutive promoter (J23102)
N = number of plasmid in a single cell
V = volume of a cell
kdegmRNA = degrading constant of sensor promoter mRNA
The aim of the equation is to know the production rate ofmRNAoxyR, and choose the proper constitutive promoter for boosting OxyR's concentration.
Equation4:
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPStrxC = promoter strength of trxC
KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant of CI
ncI = Hill coefficient of CI
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter Mrna
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how trxC promoter strength (in PoPS) influences the production of LuxI.
Equation5:
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPStrxC = promoter strength of trxC
KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant of CI
ncI = Hill coefficient of CI
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter Mrna
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how trxC promoter strength (in PoPS) influences the production of LuxR.
Equation6:
KdLuxRAHL = dissociation constant ofLuxRAHL
nLuxRAHL = Hill coefficient of LuxRAHL
KdcI = dissociation constant of CI
ncI = Hill coefficient of CI
PoPSLuxcI = promoter strength of LuxcI hybrid promoter
kdegmRNA = degrading constant of sensor promoter Mrna
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of kill protein production rate and when it can reach the level ofthe desired concentration.
Equation7:
KdROSoxyR = dissociation constant ofROSoxyR
nROSoxyR = Hill coefficient of ROSoxyR
PoPStrxC = promoter strength of trxC
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
kdegmRNA = degrading constant of sensor promoter Mrna
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of T7 polymerase production rate and when it can reach the level to translate enough T7 polymerase.
Equation8:
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
KdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of enzyme PDC production rate and when it can reach the level to translate enough PDC.
Equation9:
KdT7 = dissociation constant of T7
NT7 = Hill coefficient of T7
PoPST7 = promoter strength of T7 promoter
kdegmRNA = degrading constant of sensor promoter mRNA
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to knowmRNA of enzyme ADH production rate and when it can reach the level to translate enough ADH.
Equation10:
RBS = binding site strength
KdegCI = degrading constant of CI
The aim of the equation is to know the production rate of CI and when it can reach the concentration to repress LuxCI hybrid promoter.
Equation11:
RBS = binding site strength
KdegLacI = degrading constant of LacI
The aim of the equation is to knowLacI production rate and when it can reach the concentration to repress promoter LacI.
'Equation12:
RBS = binding site strength
KAHL = LuxI to AHL rate constant
KdegLuxI = degrading constant of LuxI
The aim of the equation is to know the production rate of LuxI concerning the LuxIAHL reaction and LuxI degrading.
'Equation13:
RBS = binding site strength
KmAHL =AHL to LuxI rate constant
KdegLuxR= degrading constant of LuxR
KonAHL = 2AHL + LuxR to (AHL)2/LuxR complex rate constant
KdegAHL = degrading constant of AHL
The aim of the equation is to know the production rate of LuxR concerning the 2AHL + LuxR to (AHL)2/LuxR reaction , AHL to LuxI reaction, and LuxR, AHL degrading.
'Equation14:
KAHL = LuxI to AHL rate constant
KonAHL = 2AHL + LuxR to (AHL)^2/LuxR complex rate constant
KoffAHL = (AHL)2/LuxR complex to 2AHL + LuxR rate constant
KdegAHL = degrading constant of AHL
The aim of the equation is to know the production rate of AHL concerning the LuxIAHL reaction, 2AHL + LuxR to (AHL)2/LuxR reaction, (AHL)2/LuxR complex to 2AHL + LuxR reaction and AHL degrading.
'Equation15:
RBS = binding site strength
KdegT7 = degrading constant of T7
The aim of the equation is to know the threshold concentration of oxyR to conquer the terminal and when T7 polymerase can reach the required concentration to activate T7 promoter.
'Equation16:
RBS = binding site strength
PoPST7 = promoter strength of T7 promoter
N = number of plasmid in a single cell
V = volume of a cell
KdegPDC = degrading constant of PDC (pyruvate decarboxylase)
The aim of the equation is to know PDC production rate and when it can reach the concentration of ethanol pathway equilibrium.
'Equation17:
RBS = binding site strength
PoPST7 = promoter strength of T7 promoter
N = number of plasmid in a single cell
V = volume of a cell
KdegADH = degrading constant of ADH (Acetaldehyde)
The aim of the equation is to know ADH production rate and when it can reach the concentration of ethanol pathway equilibrium.
'Equation18:
KAHL = LuxI to AHL rate constant
KonAHL = 2AHL + LuxR to (AHL)2/LuxR complex rate constant
KoffAHL = (AHL)2/LuxR complex to 2AHL + LuxR rate constant
KdegAHL = degrading constant of AHL
The aim of the equation is to know the production rate of AHL concerning the LuxIAHL reaction, 2AHL + LuxR to (AHL)2/LuxR reaction, (AHL)2/LuxR complex to 2AHL + LuxR reaction and AHL degrading.
'Equation19:
RBS = binding site strength
Kdegkill = degrading constant of kill
The aim of the equation is to knowkiller protein production rate and when it can reach the effective concentration to killNosema.
'Equation20:
Kpyruvateacetaldehyde = pyruvate→acetaldehyde reaction rate constant
Kacetaldehydeethanol = acetaldehyde→ethanol reaction rate constant
Km = ethanol→acetaldehyde reaction rate constant
KdADH = dissociation constant of ADH
'The aim of the equation is to know ethanol production rate and when it can reach the concentration to kill the spores of Nosema Ceranae.
Explanation:
In this equation, PoPSpLac represents the promoter strength of LacI promoter, which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription; \frac{1-[LacI]^nLacI }{ KdLacI^nLacI+[LacI]^nLacI } represents the hill effect of repressor LacI to LacI promoter. Because LacI is a repressor, the numerator is {1-[LacI]^nLacI } . For the section of the equation, \frac{1-[LacI]^nLacI}{ KdLacI^nLacI+ [LacI]^nLacI} \times PoPSconstitutive\times\frac{N}{V}represents the synthesizing rate of mRNACI under the influence of LacI and LacI promoter; -{kdegmRNA}\times {[mRNACI]} represents the degrading rate of mRNA
In this equation, RBS represents ribosome binding site strength, which is the affinity of ribosome to the starting site of mRNA.
For the section of the equation, RBS\times [mRNAcI] represents the synthesizing rate of CI; -{kdegCI}\times {[CI]} represents the degrading rate of CI.
Since LuxCI hybrid promoter is CI dominant, the presence of CI block the promoter and kill protein is not produced. This situation happens when there is no Nosema in bees.
In this equation, PoPStrxC represents the promoter strength of promotertrxC, which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription; \frac{1-[ROSoxyR]^nROSoxyR }{ KdROSoxyR ^nROSoxyR + [ROSoxyR]^nROSoxyR} represents the hill effect of activator ROSoxyR to trxCpromoter.
For the section of the equation, \frac{1-[ROSoxyR]^nROSoxyR }{ KdROSoxyR ^nROSoxyR + [ROSoxyR]^nROSoxyR} \times {PoPStrxC}\times\frac{N}{V} represents the synthesizing rate of mRNALacI under the influence of activator ROSoxyR complex and trxC promoter; -kdegmRNA\times [mRNAlacI] represents the degrading rate of mRNALacI.
In this equation, RBS represents ribosome binding site strength, which is the affinity of ribosome to the starting site of mRNA. For the section of the equation, RBS\times [mRNALacI] represents the synthesizing rate of LacI; -KdegCI\times [LacI] represents the degrading rate of LacI .
LacI will repress pLac promoter, which leads to no production of CI. Without CI, LuxCI hybrid promoter will no longer be repressed, and kill protein can thus be generated. This situation happens when Nosema exists and the sensor promoter is turned on.
In this equation, PoPSconstitutive represents the promoter strength of the constitutive promoter J23102, which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription.
For the section of the equation, PoPSconstitutive\times\frac{N}{V} represents the synthesizing rate of mRNA"oxyR" ; -KdegmRNA\times [mRNAOxyR] represents the degrading rate of mRNA"oxyR" .
In this equation, PoPStrxC,PoPSLuxcI represents the promoter strength of promoter trxC and LuxcI hybrid promoter, which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription; \frac{1-[ROSoxyR]^n ROSoxyR }{ Kd ROSoxyR ^n ROSoxyR + [ROSoxyR]^n ROSoxyR I} represents the hill effect of activator ROSoxyR complex to trxCpromoter; \frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL} represents the hill effect of activator LuxRAHL to LuxcI hybrid promoter; \frac{1-[CI leak]^nCI}{ KdCI^nCI+[CI]^nCI} represents the hill effect of repressor CI, while frac{1-{1-[CI leak]^nCI}}{ KdCI^nCI+[CI]^nCI} takes CI leaking into consideration (promoter pLac will not close thoroughly even though CI exists).
For the section of the equation, \frac{1-[ROSoxyR]^n ROSoxyR }{ Kd ROSoxyR ^n ROSoxyR + [ROSoxyR]^n ROSoxyR I} \times PoPStrxC\times\frac{N}{V} represents the synthesizing rate of mRNALuxI under the influence of activator ROSoxyR complex and trxC promoter; \frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL}\times\frac{1-{1-[CI leak]^nCI}}{ KdCI^nCI+[CI]^nCI}\times PoPSLuxCI\times\frac{N}{V} represents the synthesizing rate of mRNALuxI under the influence of LuxRAHL complex, CI repression and leaking, and LuxcI hybrid promoter; -kdegmRNA\times [mRNAluxI]represents the degrading rate of mRNAluxI.
In this equation, RBS represents ribosome binding site strength, which is the affinity of ribosome to the starting site of mRNA; KAHL represents the rate constant of AHL to LuxI. For the section of the equation, {RBS}\times {[mRNALuxI]} represents the synthesizing rate of LuxI; KAHL[LuxI] represents the transforming rate of AHL to LuxI; -KdegLuxI\times [LuxI] represents the degrading rate of LuxI.
LacI will repress pLac promoter, which leads to no production of CI. Without CI, LuxCI hybrid promoter will no longer be repressed, and kill protein can thus be generated. This situation happens when Nosema exist, and when the sensor promoter is turned on.
In this equation, PoPStrxC,PoPSLuxcI represents the promoter strength of promoter trxC and LuxcI hybrid promoter, which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription; \frac{1-[ROSoxyR]^n ROSoxyR }{ Kd ROSoxyR ^n ROSoxyR + [ROSoxyR]^n ROSoxyR I} represents the hill effect of activator ROSoxyR complex to trxCpromoter; \frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL} represents the hill effect of activator LuxRAHL to LuxcI hybrid promoter; \ frac{1-[CI leak]^nCI}{ KdCI^nCI+[CI]^nCI} represents the hill effect of repressor CI, while frac{1-{1-[CI leak]^nCI}}{ KdCI^nCI+[CI]^nCI} takes CI leaking into consideration (promoter pLac will not close thoroughly even though CI exists).
For the section of the equation, \frac{1-[ROSoxyR]^n ROSoxyR }{ Kd ROSoxyR ^n ROSoxyR + [ROSoxyR]^n ROSoxyR I} \times PoPStrxC\times\frac{N}{V} represents the synthesizing rate of mRNALuxR under the influence of activator ROSoxyR complex and trxC promoter; \frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL}\times\frac{1-{1-[CI leak]^nCI}}{ KdCI^nCI+[CI]^nCI}\times PoPSLuxCI\times\frac{N}{V} represents the synthesizing rate of mRNALuxR under the influence of LuxRAHL complex, CI repression and leaking, and LuxcI hybrid promoter; -kdegmRNA\times [mRNAluxI]represents the degrading rate of mRNAluxR.
In this equation, RBS represents ribosome binding site strength, which is the affinity of ribosome to the starting site of mRNA; KmAHL represents the rate constant of AHLLuxI; KonAHL represents the rate constant of 2AHL+LuxR to AHL2LuxR complex.
For the section of the equation, { RBS}\times{ [mRNALuxR]} represents the synthesizing rate of LuxR; { -KmAHL}\times{ [AHL]} represents the transforming rate of AHL to LuxI; {-KdegLuxR}\times {[LuxR]} represents the degrading rate of LuxR; { -KonAHL}\times{ [AHL]^2}\times{ [LuxR]} represents the transforming rate of 2AHL+LuxR to AHL2LuxR complex; {-KdegLuxI}\times {[AHL]} represents the degrading rate of AHL.
In this equation, KAHL represents the rate constant of AHL to LuxI; KoffAHL represents the rate constant of AHL2LuxR complex to 2AHL+ LuxR, while KonAHL represents the reverse rate constant.
For the section of the equation, {KAHL}\times {[LuxI]} represents the transforming rate of LuxI to AHL; {2}\times {KoffAHL}\times{ [AHLLuxR]} represents the transforming rate of AHL2LuxR complex to AHL and LuxR, the constant 2 means that each complex turns into two AHL; -{2}\times {KonAHL}\times {[AHL]^2}\times {[LuxR]} represents the transforming rate of 2AHL+LuxR to AHL2LuxR complex; {-KdegLuxI}\times {[LuxI]} represents the degrading rate of LuxI.
Since LuxI will transform to AHL, and 2AHL will combine to LuxR to form AHL2LuxR complex. Because AHL2LuxR will then bind to LuxCI hybrid promoter and activate kill gene, LuxR gene, and LuxI gene. In this way, kill protein will kill Nosema, while LuxR, LuxI will trigger the positive feedback and enhance the production of kill protein.
After trxC promoter is turned on by ROSoxyR complex and the downstream genes – LacI, LuxI, and LuxR starts to produce, which blocks the CI promoter and turns on the LuxCI hybrid promoter. After that, the gene encoded with kill protein downstream LuxCI hybrid promoter also begins to transcribe.
This equation shows production rate of mRNAkill. PoPSLuxcI represents the promoter strength of promoter trxC and LuxcI hybrid promoter, which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription; \frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL} represents the hill effect of activator LuxRAHL to LuxcI hybrid promoter; \ frac{1-[CI leak]^nCI}{ KdCI^nCI+[CI]^nCI} represents the hill effect of repressor CI, while frac{1-{1-[CI leak]^nCI}}{ KdCI^nCI+[CI]^nCI} takes CI leaking into consideration (promoter pLac will not close thoroughly even though CI exists).
For the section of the equation, \frac{1-[ LuxRAHL]^nLuxRAHL}{ KdLuxRAHL ^nLuxRAHL + [LuxRAHL]^nLuxRAHL}\times \frac {1-[CI leak]^nCI}{ KdCI^nCI+[CI]^nCI}\times {PoPSLuxcI} \times \frac{N}{V}
represents the synthesizing rate of mRNAkill under the influence of LuxRAHLCI repression and leaking, and LuxcI hybrid promoter; -kdegmRNA\times [mRNALuxR] represents the degrading rate of mRNAkill.
As for explanations of equation7, 8, 9, 15, 16, 17, 20, please see the ethanol wiki for more information.
Results:
After Nosema invasion (t=0), concentration of kill protein will reach its constant level, which is 6.5\times 10^(-9)M. The effective concentration of kill protein to kill Nosema is 6.7\times 10^(-10)M, which is near the level we have modeled. Besides, the time to reach this concentration is 24 hours after Nosema invasion. Since three days after Nosema invasion, bees will start to spread Nosema, the constant level of kill protein (which is a little higher than the effective one)and the time it needs to reach this level (which is a little bit shorter than )shows that Bee. Coli can save the bees timely.
Similarly, the effective concentration of ethanol to kill bees is 2.4\times 10^(-6)M and the time to reach this concentration is 72 hours after Nosema invasion. Since kill protein will kick on two days after Nosema invasion, the open time of ethanol (four days after Nosema invasion) is what we desired. That is, because ethanol is the last end of preventing Nosema from spreading (it kills the infected bees as well), we want the open time of ethanol to be later than kill protein open time.
The benefits of our circuit are that the dissociation constant of T7 (KdT7) is small, which means it is sensitive (will swiftly open right after T7 polymerase reaches threshold) and that we use positive feedback to attain the goal of open this device like a switch.
Parameters:
| Model | Parameter | Description | Value | Unit | Reference |
|---|---|---|---|---|---|
| Sensor | KdegOxyR | OxyR degrading rate | 107 | M-1 x min-1 | Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol—disulfide status |
| KdOxyR* | Dissociation constant of OxyR* | 10-7.33 | M | ||
| KOxyR | OxyR producing rate constant | 5.012 x 1014 | X | ||
| nOxyR* | Hill coefficient of OxyR* | Ranging from 0.75~3.5,depends on what kind of ROS it react with | X | OxyR: A Molecular Code for Redox-Related Signaling | |
| N | Copy number | 887(cells containing 25% plasmid bearing cells) | Single piece |
| |
| Ethanol
| pyruvate | Initial concentration of pyruvate in MG1655 | 1.18 x 2 | g/L | Expression of pyruvate carboxylase enhances succinate production in Escherichia coli without affecting glucose uptake |
| nPDC | Hill coefficient of PDC | 2.1 | - | Purification, characterization and cDNA sequencing of pyruvate decarboxylase Zygosaccharomyces biporus | |
| nADH | Hill coefficient of ADH | at high pH values, 30◦C, n=1; at low temperature, n=3 | - | Evidence for co-operativity in coenzyme binding to tetrameric Sulfolobus
solfataricus alcohol dehydrogenase and its structural basis: fluorescence, kinetic and structural studies of the wild-type enzyme and non-co-operative N249Y mutant | |
| SEIR(exponential)
| b | Infection rate constant of Nosema ceranae to the suspected | 24/75 | Period(days)-1 | |
| r1 | Infection rate constant of K12 to the suspected | 3/20 | Period(days)-1 | 1. Environment protection administration executive yuan of R.O.C Medical bacteriology of J.A.T | |
| r2 | Infection rate constant of K12 to the latent | 3/20 | Period(days)-1 | 2. Environment protection administration executive yuan of R.O.C. 3. Medical bacteriology of J.A.T | |
| e | rate of the latent turns infectious | 1/4 | Period(days)-1 | ||
| u | Death rate of the infected | 1/8 | Period(days)-1 | ||
| k | Rate of intaking capsule | 24/11 | Period(days)-1 | ||
| SEIR(exponential&linear) | S | x(1) | Amount of total population | ||
| E | x(2) | Amount of suspected individuals | |||
| I | x(3) | Amount of individuals in the latent period | |||
| R | x(4) | Amount of infected individuals |
 "
"