Team:Bielefeld-Germany/Biosafety/Biosafety Strain
From 2013.igem.org
| Line 82: | Line 82: | ||
<br> | <br> | ||
[[File:IGEM Bielefeld 2013 Biosafety gram- ohnehintergrund 2.png|400px|thumb|center|'''Figure 2:''' The cell wall of a gram-negative bacteria is characterized by a thin peptidoglycan-layer and a second membran. ]] | [[File:IGEM Bielefeld 2013 Biosafety gram- ohnehintergrund 2.png|400px|thumb|center|'''Figure 2:''' The cell wall of a gram-negative bacteria is characterized by a thin peptidoglycan-layer and a second membran. ]] | ||
| - | [[File:IGEM Bielefeld 2013 Biosafety gram+ ohnehintergrund 2.png|400px|thumb|center|'''Figure 3:'''' The cell wall of a gram-positive bacteria instead is characterized by a thick peptidoglycan layer and only one membrane]] | + | [[File:IGEM Bielefeld 2013 Biosafety gram+ ohnehintergrund 2 verbessert.png|400px|thumb|center|'''Figure 3:'''' The cell wall of a gram-positive bacteria instead is characterized by a thick peptidoglycan layer and only one membrane]] |
<br> | <br> | ||
<p align="justify"> | <p align="justify"> | ||
Revision as of 02:21, 5 October 2013
Safety Strain
Overview
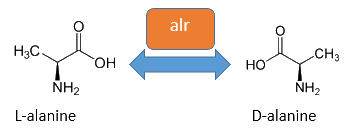
The basis for our Safety-System is an auxotrophic safety strain combined with a RNase from Bacillus amyloliquefaciens as a degradation part. This K12 descended Safety-Strain the constitutive Alanine-Racemase (alr) and the catabolic Alanine-Racemase (dadX) are deleted. As the Alanine-Racemase catalyses the reversible isomerization from L-alanine into the enantiomer D-alanine our safety strain is no longer able to synthesis the essential amino acid D-alanine. D-alanine is an essential molecule for the gram-negative bacteria E. coli, because it is responsible for the cross-linkage of the peptidoglycan layer, so that a lack of D-alanine inhibits cell division and leads to cell lysis in growing bacteria. This effect is comparable to bacteristatic characteristic known from beta-lactam-antbiotics.
An important parameter, when expressing toxic gene products, like the Rnase Ba (barnase), is the maintenance of the plasmid stability. The strict dependance on D-alanine is used to increase the plasmid stability, when the plasmid contains the coding sequence for the Alanine-Racemase (alr) and no D-alanine is supplemented. Therefore, the plasmid is necessary for the growing, because otherwise the bacteria are not able to grow. So by using this system, it is more certain that the plasmid with the toxic gene product is kept by the cell. Additionally this causes a double kill-switch system, because now not only the barnase would imply cell death, but also the loss of the Alanine-Racemase (alr) encoded on the plasmid.
Theory
The cell wall is essential for every living bacteria, because it divides the cytoplasm from the environment. In fact, the life of the bacteria is only possible because of this separation. On the one hand the cell wall confers stability and structure and on the other hand it enables the osmotic pressure. Further the cell wall regulates the transport of molecules across the cell wall.
The composition of the cell wall differs between the bacteria, so that the bacteria generally are divided into gram-negative Bacteria, for example Escherichia coli and gram-positive Bacteria for example Bacillus subtillis. Gram-negative bacteria therefore are characterized by an inner plasma membrane, a thin peptidoglycan layer, periplasmatic spaces an the outer membrane. While gram-positive bacteria in contrast poses only one periplasmatic space but a thicker peptidoglycan layer (see figure below).
Hence, it becomes clear that the peptidoglycan layer is an interesting approach to control bacterial growth. Peptidoglycan itself is a polymer consisting of a linear chain of polysaccharides and short peptides. The polysaccharides component are of alternating residues of beta-(1,4) linked N-acetylglucosamine and N-acetylmuramic acid and they are cross-linked in E. coli by a tetra-peptide of L-Alanine, D-glutamic acid, meso-diaminopimelic acid and final D-alanine. The cross-linkage is thereby realized by a transpeptid-linkage of meso-diaminopimelic acid and D-alanine.(Cava et al., 2011)
This points out that D-Alanin is an essential component of the bacterial peptidoglycan layer. In conclusion a lack of D-Alanin would have a bacteriostatic characteristic, comparable to beta-lactam-antibiotic, because it inhibits the building of the peptdiglycan-linkage. Hence, the bacteria without D-Alanine could not divide, because they will lyse otherwise.
The accumulation of D-alanine in E. coli can be catalyzed by an Alanine-Racemase (EC 5.1.1.1). This enzyme enables the reversible reaction from L-alanine into the enantiomer D-alanine. For this reaction the cofactor pyridoxal-5'-phosphate (PLP) is also needed. E. coli exists of two Alanine-Racemases. One Alanine-Racemase (alr) is constitutive expressed and therefore normally responsible for the accumulation of D-Alanine, while the other Alanin-Racemase (dadX) is under control of the dad-Operon and only transcripted in deficient situations (Walsh et al., 1989).
The deletion of the constitutive Alanin-Racemase (alr) and the catabolic Alanine-Racemase (dadX) will lead to a strict dependence of D-Alanine, so that the bacteria with this mutations are only able to grow on media with D-alanine supplemention or by complementation of the Alanin-Racemase on a separate plasmid. As shown in the figure below the D-Alanin-Auxotrophic mutant shows a strict dependence on D-Alanine.
Genetic Approach
Deletion of the Alanine-Racemases
For the construction of our Biosafety-Chassis we deleted both Alanine-Racemases via homologous recombination with the pKO4 (a derivation from the pKO3 containing the coding sequence of the lacZ-alpha Fragment) to establish a D-alanine-auxotrophic mutant. For the deletion of the Alanine-Racemase we thereby used the following primers.
The primers alr_Ec_d1 and alr_Ec_d2 will amplify an about 600 bp long homologous fragment upstream of the coding sequence of the Alanine-Racemase, while the primers alr_Ec_d3 and alr_Ec_d4 will yield an about 600 bp long homologous fragment downstream of the corresponding Alanine-Racemase. This two deletion-sides can be ligated by Gibson-Assembly or alternative classically by gene SOEing (Gene splicing by overlap extension, Horton et al., 1990). Afterwards the deletion-sides of the Alanine-Racemase were cloned into to vector by using the bold primer-overhang (see box below).
By following the protocol for the double-cross over using the heat-sensitive RepA protein and the sacB gene from the vector [http://arep.med.harvard.edu/labgc/pko3.html pKO3], the two Alanine-Racemases could be successfully deleted (Link et al., 1990).
Primer for the complete deletion of the constitutive Alanine-Racemase (alr) and the catabolic Alanine-Racemase (dadX) in E. coli
Primer: alr_Ec_d1 (39 bp)
5'-TAGCTCACTCATTAGGCACCCAGCTCGATGACGAAGACT-3'
Primer: alr_Ec_d2 (20 bp)
5'-GCCGCTTGCATTTGTGTTCC-3'
Primer: alr_Ec_d3 (40 bp)
5'-GGAACACAAATGCAAGCGGCTTGATTGTCTGTGCCGGATG-3'
Primer: alr_Ec_d4 (40 bp)
5'-GCTTTCTACGTGTTCCGCTTCCGGGAAGTAGCGTTTCAGG-3'
Primer: dadX_Ec_d1 (39 bp)
5'-TAGCTCACTCATTAGGCACCTGAAGTGTGGCGATGAAGT-3'
Primer:dadX_Ec_d2 (20 bp)
5'-GGGTCATCTCGTTTCCTTAG-3'
Primer: dadX_Ec_d3 (40 bp)
5'-CTAAGGAAACGAGATGACCCACTTGTTGTAAGCCGGATCG-3'
Primer: dadX_Ec_d4 (40 bp)
5'-GCTTTCTACGTGTTCCGCTTCGAAGCCAGCGCCAAATATG-3'
To verify the deletion of the Alanine-Racemases in E. coli we designed some additional primers, which anneal outside the deletion region so that a successful deletion can be seen by a 1370 bp (alr) or a 1312 bp (dadX) amplified DNA-fragment, while the wild type shows typically a DNA-fragment with a length of 2439 bp (alr) or 2375 bp (dadX). The primers for the control are listed in the box below.
Primer for the control of the successful Deletion of the constitutive Alanine-Racemase (alr) and the catabolic Alanine-Racemase (dadX) in E. coli
Primer: alr_Ec_control1 (20 bp)
5'-GCTGGAGATGCCATCAGAAC-3'
Primer: alr_Ec_control2 (20 bp)
5'-CCGGCGAATATTGCTACGTG-3'
Primer: dadX_Ec_control1 (20 bp)
5'-GCTTTAATACGCCCGTTGAC-3'
Primer: dadX_Ec_control2 (20 bp)
5'-CTGGATCAACGCTTCTTTGG-3'
Deletion of araC
In addition to the deletion of the Alanine-Racemaes we deleted the coding sequence for araC in the Safety-Strain. As the AraC protein is the repressor of the pBAD (Arabinose) promoter this mutation effects only the Biosafety-System araCactive, but is very useful because of two reasons: On the one hand the repressor AraC works also as an activator, so that there is no basal transcription behind the pBAD promoter in the strain E. coli K-12 Δalr ΔdadX ΔaraC as verified. This opens the possibility to transform the Biosafety-System araCative with the barnase as the toxic gene product in the bacteria without triggering cell death. On the other hand the pBAD promoter is possible too tight regulated by the repressor AraC, that a genomic deletion is necessary for the expression of the toxic gene barnase, when there is still a basal transcription of the araC gene from the plasmid.
For the deletion of the repressor AraC we therefore used the same methods as for the deletion of the Alanine-Racemases in E. coli. The primers araC_d1 and araC_d2 will amplify analogous of about 600 bp long homologous fragment upstream of the coding sequence of the coding sequence for araC, while the primers araC_d3 and araC_d4 will perform an about 600 bp long homologous fragment downstream of the araC. This two deletion-sides can be ligated analog by Gibson-Assembly or gene SOEing (Gene splicing overlap Extension, Horton et al., 1990) and then cloned into to gene-targeting vector [http://arep.med.harvard.edu/labgc/pko3.html pKO3] by using the bold primer-overhang (see box below). By following the protocol for the double-cross over by using the heat-sensitive RepA protein and the sacB from the vector pKO3, the repressor AraC could be successfully deleted (Link et al., 1990).
Primer for the complete Deletion of the repressor araC for the arabinose promoter pBAD in E. coli
Primer: araC_d1 (40 bp)
5'-TAGCTCACTCATTAGGCACCCCGGCAGGAATATCGATCGC-3'
Primer: araC_d2 (40 bp)
5'-CTTCTCTGAATGGTGGGAGTGTCATAATTGGTAACGAATC-3'
Primer: araC_d3 (40 bp)
5'-GATTCGTTACCAATTATGACACTCCCACCATTCAGAGAAG-3'
Primer: araC_d4 (40 bp)
5'-GCTTTCTACGTGTTCCGCTTAACGCCAATCCCGACCACAG-3'
To verify the deletion of the repressor AraC in E. coli we designed some primer, which anneal outside the deletion region so that a successful deletion can be seen by a 1404 bp amplified DNA-fragment, while the wild type shows typically a DNA-fragment with a length of 2268 bp. The primers for the control are listed in the box below.
Primer for the control of the successful Deletion of repressor araC in E. coli
Primer: araC_control1 (20 bp)
5'-CTTTACCGCTGCGCCATAAC-3'
Primer: araC_control2 (20 bp)
5'-AACCGCAGTGTGGTCTTTCC-3'
Results
Characterization of the D-alanine auxotrophy
The deletion of the Alanin-Racemases and araC in E. coli was not possible in the common used strains like JM109, Top10 or KRX, but in the wild type strain K-12. This is may due to the RecA1-mutations in this strains, which guarantees a better plasmid maintenance because of defect rekombinase.
To avoid a second recombination of the Alanine-Racemase (alr) from the plasmid with the genome, the whole coding sequence was deleted in the genome and the characterization of the Alanine-Racemase was performed with the antibiotic chlormaphenicol. For the complementation the Alanine-Racemase (alr) was brought under the control of the ptac promoter. The ptac promoter is a fusion promoter of the -35-region of the trp promoter and the -10-region the lac promoter, so that there only slight repression and the expression of the Alanine-Racemase is highly activated (De Boer et al., 1983). Therefore an induction with IPTG was not necessary on M9, but surprisingly it was essential on LB-agar.
The deletion of the constitutive Alanine-Racemase (alr) and the catabolic Alanine-Racemase (dadX) in E. coli leads to a strict dependance on the amino acid D-alanine, as aspected. As shown in the figure below the bacteria with this deletions are not any more able to grow on normal M9-media without D-alanine supplementation (purple curve), whereas the wild type does (red curve). The auxotrophic Safety-Strain grows only on media with D-alanine (5 mM) supplemented (blue curve) or by a complementation of the Alanine-Racemase via plasmid. Further it can be seen, that the auxotrophic mutant K-12 ∆alr ∆dadX grows slightly slower, than the wild type K-12. In contrast the bacteria containing the Alanine-Racemase (alr) on the plasmid <bbpart>BBa_K1172902</bbpart> does hardly show a disadvantage in the cell division compared to the wild type.
Characterization of the araC-deletion
The deletion of the araC gene was added to the D-alanine auxotrophic Biosafety-Strain resulting in the E. coli strain K-12 ∆alr ∆dadX ∆araC. This Strain is in addition to the D-alanine auxotrophy, as aspected, not any more able to use the pentose L-arabinose as carbon source. This opens the possibility to maintain a plasmid with a toxic gene behind the pBAD promoter without causing cell death. This huge advantage is caused by the fact that the araC protein functions as repressor and activator of the arabinose operon pBAD simultaneously. Lacking this activator the transcription of the genes behind this promoter is shutdown completely.
To verify if the pBAD promoter is really shutdown the Biosafety-Strain K-12 ∆alr ∆dadX ∆araC were cultivated on M9-minimal media containing only L-arabinose as carbon source. As shown in the figure (6) below without the araC gene there is not even a slight basal transcription (blue curve). The Biosafety-Strain can not grow on L-arabinose, where as the positive control (red curve) does. In addition the growth of the wild-type on L-arabinose was compared to D-glucose (black curve). The faster growth of E. coli on D-glucose in contrast to L-arabinose explains why the pBAD operon is naturally shut down. Because D-glucose is more efficient to metabolize, a tight regulation of the arabinose catabolism is needed to save the energy resource of the cell. But in synthetic biology this regulon can be used for different approaches. The Biosafety-System araCtive is only one interessting application.

References
- Cava F, Lam H, de Pedro MA, Waldor MK (2011) Emerging knowledge of regulatory roles of d-amino acids in bacteria [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3037491/pdf/18_2010_Article_571.pdf|Cell and Molecular Life Sciences 68: 817 - 831.]
- De Boer Hermann, Comstock J. Lisa and Vasser Mark (1983) The tac promoter: a functional hybrid derived from the trp and lac promoters. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC393301/pdf/pnas00627-0036.pdf|Proceedings of the National Academy of Science of the United States of America 80: 21 - 25.]
- Horton R., Cai Z., Ho S. and Pease L. (1990) Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction [http://www.ncbi.nlm.nih.gov/pubmed/2357375|BioTechniques 8: 528 - 535.]
- Link, A.J., Phillips, D. and Church, G.M. (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC179534/pdf/1796228.pdf|Journal of Bacteriology 179: 6228-6237.]
- Walsh, Christopher (1989) Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. [http://www.jbc.org/content/264/5/2393.long|Journal of biological chemistry 264: 2393 - 2396.]
 "
"