Team:Bielefeld-Germany/Labjournal/June
From 2013.igem.org
(Difference between revisions)
| Line 70: | Line 70: | ||
====Mediators==== | ====Mediators==== | ||
*Glycerol dehydrogenase | *Glycerol dehydrogenase | ||
| - | **Isolation of shipping vector pSB1C3 out of 2013 Distribution Kit Plate 5 Well 3A with insert Part RFP (J04450) for better transformation characterization (Distribution Kit BioBrick isolation). | + | **Isolation of shipping vector pSB1C3 out of 2013 Distribution Kit Plate 5 Well 3A with insert Part RFP (<bbpart>J04450</bbpart>) for better transformation characterization ([http://parts.igem.org/Help:2013_DNA_Distribution Distribution Kit BioBrick isolation]). |
| - | **Transformation of <partinfo>BBa_J04450</partinfo> into ''Escherichia coli'' KRX strain. | + | **[[Team:Bielefeld-Germany/Labjournal/Molecular# Transformation via electroporation| Transformation]] of <partinfo>BBa_J04450</partinfo> into ''Escherichia coli'' KRX strain. |
| - | **Plasmid isolation of <partinfo>BBa_J04450</partinfo>. | + | **[[Team:Bielefeld-Germany/Labjournal/Molecular#Used Kits | Plasmid isolation]] of <partinfo>BBa_J04450</partinfo>. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| Line 93: | Line 78: | ||
===Week 7=== | ===Week 7=== | ||
| - | |||
| - | |||
| - | |||
====MFC==== | ====MFC==== | ||
| Line 123: | Line 105: | ||
**Lane2: 4-2106-451: 7.4 ng/µl | **Lane2: 4-2106-451: 7.4 ng/µl | ||
**Lane5: 4-2106-451: 8.5 ng/µl | **Lane5: 4-2106-451: 8.5 ng/µl | ||
| - | |||
| - | |||
| - | |||
| - | |||
====Porines==== | ====Porines==== | ||
| - | + | *Starting first cultivation of ''Pseudomonas fluorescens'' strain for [[Team:Bielefeld-Germany/Labjournal/Molecular#Whole Genome Isolation | complete genome isolation]]. | |
| - | + | *Successful [[Team:Bielefeld-Germany/Labjournal/Molecular#Whole Genome Isolation | genome isolation]] of ''Pseudomonas fluorescens''. | |
| - | *Starting first cultivation of ''Pseudomonas fluorescens'' strain for | + | |
| - | *Successful [ | + | |
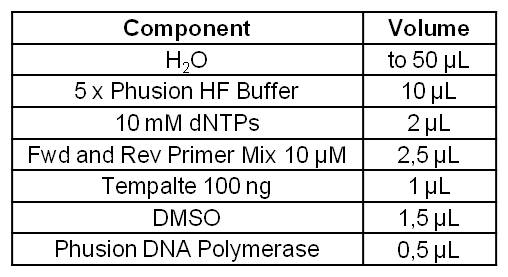
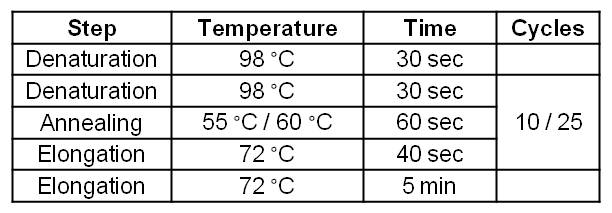
*Successful PCR with Forward and Reverse Primer OprF on the OprF gene of ''Pseudomonas fluorescens'' strain. | *Successful PCR with Forward and Reverse Primer OprF on the OprF gene of ''Pseudomonas fluorescens'' strain. | ||
[[Image:IGEM_Bielefeld_Standard_Phusion_PCR_Master_MixLRO.jpg|300px|thumb|left|<p align="justify"> '''Table 1: Standard Phusion PCR Master Mix. '''</p>]] | [[Image:IGEM_Bielefeld_Standard_Phusion_PCR_Master_MixLRO.jpg|300px|thumb|left|<p align="justify"> '''Table 1: Standard Phusion PCR Master Mix. '''</p>]] | ||
| - | |||
[[Image:IGEM_Bielefeld_Standard_Phu_PCR_GldA_OprF.jpg|300px|thumb|center| <p align="justify">'''Table 2: Two step standard Phusion PCR programm for GldA amplification. '''</p>]] | [[Image:IGEM_Bielefeld_Standard_Phu_PCR_GldA_OprF.jpg|300px|thumb|center| <p align="justify">'''Table 2: Two step standard Phusion PCR programm for GldA amplification. '''</p>]] | ||
| - | + | *OprF PCR product was isolated by Agarose gel electrophorese and [[Team:Bielefeld-Germany/Labjournal/Molecular#Used Kits | purificated]]. | |
| - | + | ||
| - | *OprF PCR product was isolated by Agarose gel electrophorese and purificated. | + | |
*Bands are at expected size of 1300 bp. | *Bands are at expected size of 1300 bp. | ||
| - | [[Image:IGEM_Bielefeld_OprF_standard_PCR.jpg|200px|thumb|left| <p align="justify">'''Figure 1: Agarosegel from PCR on the OprF gene of ''Pseudomonas fluorescens'' strain with forward and reverse primer OprF. For Ladder we used GeneRuler™ 1 kb DNA Ladder fromThermo Scientific. '''</p>]] | + | [[Image:IGEM_Bielefeld_OprF_standard_PCR.jpg|200px|thumb|left| <p align="justify">'''Figure 1: Agarosegel from PCR on the OprF gene of ''Pseudomonas fluorescens'' strain with forward and reverse primer OprF. For Ladder we used [http://www.thermoscientificbio.com/nucleic-acid-electrophoresis/generuler-1-kb-dna-ladder-ready-to-use-250-to-10000-bp GeneRuler™ 1 kb DNA Ladder fromThermo Scientific]. '''</p>]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| Line 168: | Line 132: | ||
===Week 8=== | ===Week 8=== | ||
| + | |||
====Organization==== | ====Organization==== | ||
| - | *Let’s go to Lyon, flights are booked for the European jamboree in Lyon from 11.-13. October 2013. | + | *Let’s go to Lyon, flights are booked for the [https://2013.igem.org/Europe/Pre-Jamboree European jamboree in Lyon] from 11.-13. October 2013. |
| - | *We will participate at | + | *We will participate at ‘[http://www.bio.nrw.de/studentconvention BioNRW pHD Student Convention]’ in Düsseldorf at 13. July. |
| + | |||
====MFC==== | ====MFC==== | ||
| Line 178: | Line 144: | ||
====Mediators==== | ====Mediators==== | ||
*Glycerol dehydrogenase | *Glycerol dehydrogenase | ||
| - | |||
**Cloning of GldA into pSB1C3 shipping vector with NEB Biobrick assembly Kit did not work as expected. | **Cloning of GldA into pSB1C3 shipping vector with NEB Biobrick assembly Kit did not work as expected. | ||
| - | **Screening of colonies with colony PCR and Plasmid restriction analysis shows religated pSB1C3 shipping vector. | + | **Screening of colonies with [[Team:Bielefeld-Germany/Labjournal/Molecular#Colony PCR | colony PCR]] and Plasmid [[Team:Bielefeld-Germany/Labjournal/Molecular#Restriction analysis | restriction analysis]] shows religated pSB1C3 shipping vector. |
**Bands are at size of 2000 bp, the length of linear pSB1C3. | **Bands are at size of 2000 bp, the length of linear pSB1C3. | ||
| - | [[Image:IGEM_Bielefeld_Religierter_pSB1C3_restriktion.jpg|200px|thumb|left| <p align="justify">'''Figure 2: Agarosegel with NEB 1 kb DNA Ladder as marker. Bands are showing restriction analysis from cloning of GldA into pSB1C3 shipping vector with NEB Biobrick assembly Kit. Assembly did not work, only one band at the size of 2000 bp showing religated pSB1C3. '''</p>]] | + | [[Image:IGEM_Bielefeld_Religierter_pSB1C3_restriktion.jpg|200px|thumb|left| <p align="justify">'''Figure 2: Agarosegel with [https://www.neb.com/products/n3232-1-kb-dna-ladder NEB 1 kb DNA Ladder] as marker. Bands are showing [[Team:Bielefeld-Germany/Labjournal/Molecular#Restriction analysis | restriction analysis]] from cloning of GldA into pSB1C3 shipping vector with NEB Biobrick assembly Kit. Assembly did not work, only one band at the size of 2000 bp showing religated pSB1C3. '''</p>]] |
| - | + | **Primerdesign for pSB1C3 according an universal usable backbone for [[Team:Bielefeld-Germany/Labjournal/Molecular# Gibson assembly | Gibson Assembly]] with Prefix and Suffix specific overlaps: | |
| - | + | ||
| - | + | ||
| - | **Primerdesign for pSB1C3 according an universal usable backbone for Gibson Assembly with Prefix and Suffix specific overlaps: | + | |
***Forward Primer pSB1C3 (23 bp): TACTAGTAGCGGCCGCTGCAGTC | ***Forward Primer pSB1C3 (23 bp): TACTAGTAGCGGCCGCTGCAGTC | ||
***Reverse Primer pSB1C3 (23 bp): CTCTAGAAGCGGCCGCGAATTCC | ***Reverse Primer pSB1C3 (23 bp): CTCTAGAAGCGGCCGCGAATTCC | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
====Porines==== | ====Porines==== | ||
*Cloning of OprF into pSB1C3 shipping vector with NEB Biobrick assembly Kit did not work as expected. | *Cloning of OprF into pSB1C3 shipping vector with NEB Biobrick assembly Kit did not work as expected. | ||
| - | *Screening of colonies with colony PCR and Plasmid restriction analysis shows religated pSB1C3 shipping vector as described for GldA cloning. | + | *Screening of colonies with [[Team:Bielefeld-Germany/Labjournal/Molecular#Colony PCR | colony PCR]] and Plasmid [[Team:Bielefeld-Germany/Labjournal/Molecular#Restriction analysis | restriction analysis]] shows religated pSB1C3 shipping vector as described for GldA cloning. |
Revision as of 23:19, 30 September 2013
June
Milestones
Week 5
Organization
- iGEM-Team Bielefeld will support the ‘CeBiTec Student Academy’ from 26.-30. August with an own experiment.
MFC
Mediators
Porines
- Primerdesign for isolation of OprF from Pseudomonas fluorescens strain, with overlaps for Biobrick Prefix and Suffix:
- Forward Primer OprF (49 bp): GAATTCGCGGCCGCTTCTAGATGAAACTGAAAAACACCTTGGGCTTTGC
- Reverse Primer OprF (51 bp): CTGCAGCGGCCGCTACTAGTATTACTTAGCTTGGGCTTCAACCTGCGCTTC
Week 6
Organization
- Thanks to NEB Biolabs for the free iGEM kit with many useful laboratory things for all German iGEM teams.
- We are working on our first press release.
- Having a short radio contribution in the Bielefeld university campus radio (radio 87.9 hertz).
MFC
Mediators
- Glycerol dehydrogenase
- Isolation of shipping vector pSB1C3 out of 2013 Distribution Kit Plate 5 Well 3A with insert Part RFP (<bbpart>J04450</bbpart>) for better transformation characterization ([http://parts.igem.org/Help:2013_DNA_Distribution Distribution Kit BioBrick isolation]).
- Transformation of <partinfo>BBa_J04450</partinfo> into Escherichia coli KRX strain.
- Plasmid isolation of <partinfo>BBa_J04450</partinfo>.
Week 7
MFC
Mediators
Cytochromes
- Cultivation of Shewanella oneidensis MR-1 in liquid LB medium at 30 °C
- Isolation of genomic DNA from S. oneidensis
- 4-2006-451: Conc.:453.1 ng/µl OD260/280: 1.92 OD260/230:2.78
- 4-2006-452: Conc.:447.1 ng/µl OD260/280: 1.92 OD260/230:2.67
- Dilution to PCR template: 4-2006-453: 5.5ng/µl
- Amplification of the mtrCAB cluster
- Gradient: 55.8-56.7-57.8-59.1-60.4-61.7-62.9-63.9
- Clear Bands at the expected 5.2kb
- PCR-CleanUp
- PCR-cleanup of No.2 and No.5 with Macherey-Nagel-Kit and NanoDrop-Measurement
- Lane2: 4-2106-451: 7.4 ng/µl
- Lane5: 4-2106-451: 8.5 ng/µl
Porines
- Starting first cultivation of Pseudomonas fluorescens strain for complete genome isolation.
- Successful genome isolation of Pseudomonas fluorescens.
- Successful PCR with Forward and Reverse Primer OprF on the OprF gene of Pseudomonas fluorescens strain.
- OprF PCR product was isolated by Agarose gel electrophorese and purificated.
- Bands are at expected size of 1300 bp.

Figure 1: Agarosegel from PCR on the OprF gene of Pseudomonas fluorescens strain with forward and reverse primer OprF. For Ladder we used [http://www.thermoscientificbio.com/nucleic-acid-electrophoresis/generuler-1-kb-dna-ladder-ready-to-use-250-to-10000-bp GeneRuler™ 1 kb DNA Ladder fromThermo Scientific].
Week 8
Organization
- Let’s go to Lyon, flights are booked for the European jamboree in Lyon from 11.-13. October 2013.
- We will participate at ‘[http://www.bio.nrw.de/studentconvention BioNRW pHD Student Convention]’ in Düsseldorf at 13. July.
MFC
Mediators
- Glycerol dehydrogenase
- Cloning of GldA into pSB1C3 shipping vector with NEB Biobrick assembly Kit did not work as expected.
- Screening of colonies with colony PCR and Plasmid restriction analysis shows religated pSB1C3 shipping vector.
- Bands are at size of 2000 bp, the length of linear pSB1C3.

Figure 2: Agarosegel with NEB 1 kb DNA Ladder as marker. Bands are showing restriction analysis from cloning of GldA into pSB1C3 shipping vector with NEB Biobrick assembly Kit. Assembly did not work, only one band at the size of 2000 bp showing religated pSB1C3.
- Primerdesign for pSB1C3 according an universal usable backbone for Gibson Assembly with Prefix and Suffix specific overlaps:
- Forward Primer pSB1C3 (23 bp): TACTAGTAGCGGCCGCTGCAGTC
- Reverse Primer pSB1C3 (23 bp): CTCTAGAAGCGGCCGCGAATTCC
- Primerdesign for pSB1C3 according an universal usable backbone for Gibson Assembly with Prefix and Suffix specific overlaps:
Porines
- Cloning of OprF into pSB1C3 shipping vector with NEB Biobrick assembly Kit did not work as expected.
- Screening of colonies with colony PCR and Plasmid restriction analysis shows religated pSB1C3 shipping vector as described for GldA cloning.
 "
"