Team:BYU Provo/Notebook/LargePhage/Springexp/Period1/Dailylog
From 2013.igem.org
(Created page with "{{TeamBYUProvo}} <br> {| width="100%" | colspan="3" | <font color="#333399" size="5" font face="Calibri"> '''Large Phage Spring Notebook'''</font> <br> <br> |- valign="top" |...") |
|||
| (10 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{| width="100%" | {| width="100%" | ||
| - | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> '''Large Phage | + | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> '''Large Phage May - June Notebook: May 1 - May 12 Daily Log'''</font> |
<br> | <br> | ||
| Line 14: | Line 14: | ||
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | + | <font size = "4"> | |
| - | : | + | : <u> '''Large Phage''' </u> </font> |
| - | : [[Team:BYU Provo/Notebook/LargePhage/ | + | : [[Team:BYU Provo/Notebook/LargePhage/Winterexp|March-April]] |
| - | : [[Team:BYU Provo/Notebook/LargePhage/ | + | : [[Team:BYU Provo/Notebook/LargePhage/Springexp|May-June]] |
| - | : [[Team:BYU Provo/Notebook/LargePhage/Fallexp| | + | : [[Team:BYU Provo/Notebook/LargePhage/Summerexp|July-August]] |
| + | |||
| + | : [[Team:BYU Provo/Notebook/LargePhage/Fallexp|September-October]] | ||
</font> | </font> | ||
| - | | style="width: | + | | style="width: 82%; background-color: transparent;"| |
| - | <font | + | <font face="Calibri" size="3"> |
| - | < | + | <font size="4"> '''5/1/13''' </font> |
| - | + | Today we scheduled out the rest of spring semester! See Below! | |
| + | Results of looking for recipes | ||
| + | M9+ media recipe (grams per liter) | ||
| + | KH,PO4 (3.0 g), Na,HPO4 (7.0 g), NaCl (0.5 g), NH4Cl (1.0 g), | ||
| + | MgSO4.7HOH (0.246 g), CaCl, (0.011 g), ferric citrate trihydrate (0.6 mg), glucose (4.0 g), and Casamino Acids (10 g; Difco). | ||
| - | + | Phage stock preparation | |
| + | http://myxo.css.msu.edu/ecoli/phagepharm.html | ||
| - | |||
| - | + | Large Phage Group Supply List: | |
| + | LB plates | ||
| + | 5 mL pipettes | ||
| + | LB top agar | ||
| + | 5-bromodeoxyuridine - 1g | ||
| - | + | M9 minimal salts media 500g - (or wherever else is cheaper) | |
| + | MgSO4 | ||
| + | CaCl | ||
| + | ferric citrate trihydrate - not sure if this is necessary...? we only need 0.6 mg per liter and we can't find where it's sold so we'll probably omit it | ||
| + | glucose | ||
| + | Casamino Acids (500 g) | ||
| - | + | Adenine (we only need 500 mg or so) | |
| + | Uracil (we only need 500 mg or so) | ||
| + | |||
| + | Magnesium chloride | ||
| + | Chloroform | ||
<br> | <br> | ||
| - | + | <font size="4"> '''5/2/13''' </font> | |
| - | + | Today we created a plan for the next little while: | |
| - | + | May 7-8 and 13-14 | |
| - | + | Prepare and mutagenize T4 phage | |
| - | + | ||
| - | + | Day 1 Start overnight growth of W3110 | |
| - | + | ||
| - | + | (http://biology.clc.uc.edu/fankhauser/labs/microbiology/Phage/Phage_Prep.htm) | |
| - | + | ||
| - | + | Day 2 before class: | |
| - | + | Grow E. coli W3110 in H-broth --find substitute-- (supplemented with 20 ug of adenine per ml) to a density of 3 x 10^8 cells/ml | |
| + | - How do we know it's at this density? | ||
| + | |||
| + | Day 2 during class: | ||
| + | Chill and pellet bacteria | ||
| + | |||
| + | Resuspend in at concentration of 1.5 x 10^8 cells/ml in M9+ medium with 5-bromodeoxyuridine (mutagen) and other supplements | ||
| + | |||
| + | Add 2 x 10^9 phage particles to 30 mL of bacteria and aerate at 37C until cleared (a few hours). Then add Chloroform. | ||
| + | - Good stopping point here | ||
| + | |||
| + | May 15 | ||
| + | Titer mutagenized phage stocks | ||
| + | |||
| + | May 17, 20 | ||
| + | Liquid culture of giant phage | ||
| + | |||
| + | Day 1 - Start overnight growth of W3110 in M9+ | ||
| + | |||
| + | Day 2 - | ||
| + | Start mid-log to to density of 4e8 cells/mL. | ||
| + | |||
| + | Add mutagenized phages to each culture (MOI of 0.5) | ||
| + | - wait 7 minutes - | ||
| + | |||
| + | Add ten times the amount of T1 (compared to T4 mutagenized phage) to lyse uninfected cells (preventing giant phage recombination and loss). | ||
| + | - T1 has short latent period | ||
| + | - Wait seven minutes | ||
| + | |||
| + | Find a way to inhibit lysis... | ||
| + | |||
| + | Pellet bacteria, resuspend in 25 mL, lyse with Chloroform, treat with DNase and Mg+ | ||
| + | Pellet debris | ||
| + | |||
| + | May 21 | ||
| + | Start centrifugation | ||
| + | |||
| + | May 22 | ||
| + | Work with small plaques | ||
| + | Mutagenize more phage | ||
| + | |||
| + | May 24 | ||
| + | Titer centrifugation fractions | ||
| + | |||
| + | May 29 – UV and plate large phage fractions | ||
| + | |||
| + | May 30 | ||
| + | Purify large phage further | ||
| + | |||
| + | First week of June – take EMs of putative giant phage | ||
| + | Second week of June – Plate out lots of samples, look for small plaques, try to purify large phage | ||
<br> | <br> | ||
| - | <font | + | <font size="4"> '''5/3/13''' </font> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | -Today we archived our phage stock of T4 Do. Dr. Grose said she couldn’t find T1 easily available so now we are going to look into alternative e.coli phage that has a short latent period. | |
<br> | <br> | ||
| + | <font size="4"> '''5/6/13''' </font> | ||
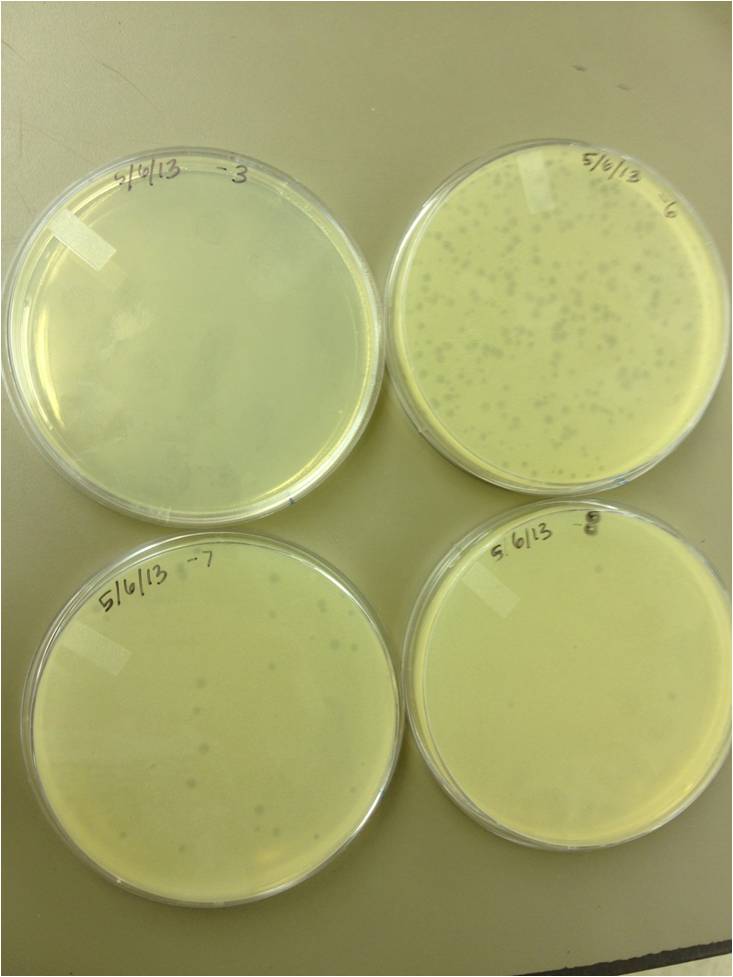
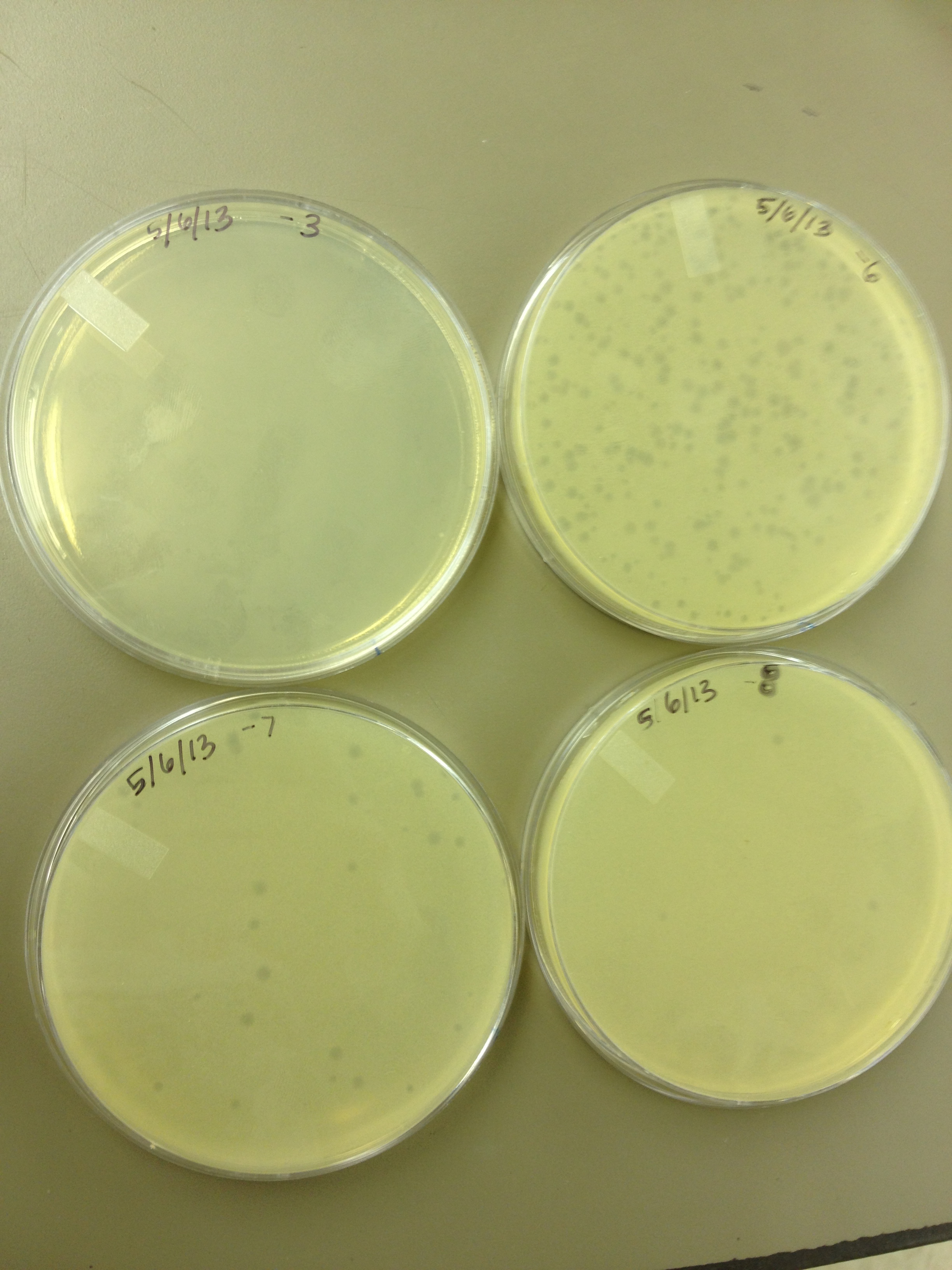
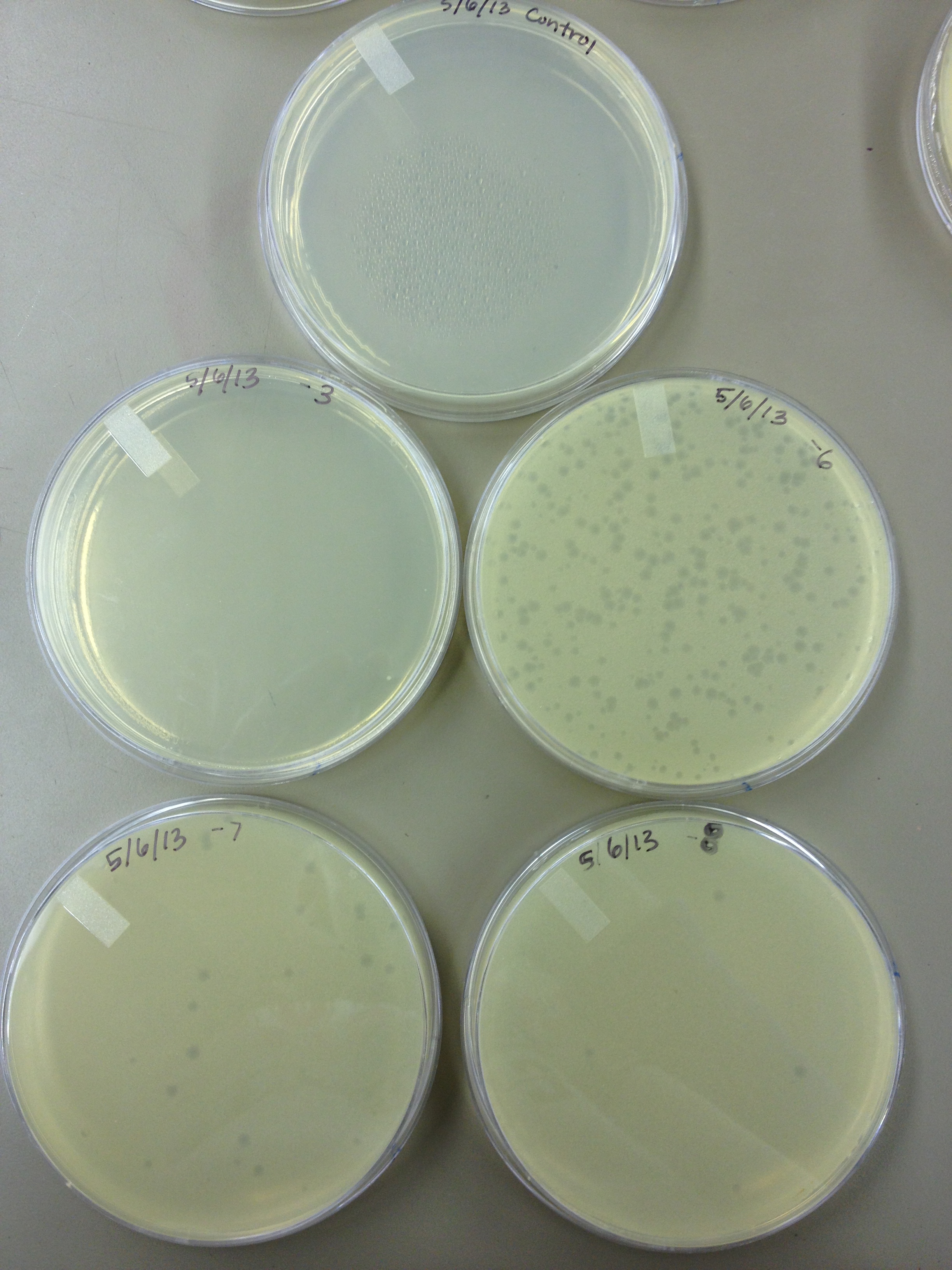
| - | + | - Today we did a phage titer of 0,-3,-6,-7,-8,-9,-10,-11 We started by putting 1 ul of phage into 1ml of bacteria, we went down from there. We infected each test tube with 10 uL from each dilution level. | |
| + | We also did a UV pre-test of sorts. We put 20ul of phage onto parafilm and exposed it to UV light in the hood of Dr. Breakwell’s lab. At 30 second intervals we took the 20 ul’s off and put it in a half ml of e.coli W3110. We tested every 30 seconds up to 3 minutes. | ||
| - | + | We also streaked out the W3110 bacteria from freezer stock onto an LB plate so we can use it to start future bacterial cultures. | |
| - | | | + | [[File:Ex.jpg|400px|center|]] |
| - | + | [[File:Ex1.jpg|400px|center|]] | |
| - | | | + | [[File:Ex2.jpg|400px|center|]] |
| - | + | [[File:Ex3.jpg|400px|center|]] | |
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | <font | + | <font size="4"> '''5/8/13''' </font> |
| - | [[ | + | -[[File:hey1.jpeg|400px|thumb|center|]] |
| + | [[File:hey2.jpeg|400px|thumb|center|]] | ||
| + | [[File:hey3.jpeg|400px|thumb|center|]] | ||
| + | [[File:hey4.jpeg|400px|thumb|center|]] | ||
| - | + | We looked at the results for the dilution series (titer) we did on the phage stock we made from the liquid culture and found that there were 5 plaques on the 10^-8 plate. | |
| - | + | We calculated pfus/mL by doing ( # of plaques ) / (dilution level x mLs infected with). Our stock is between 3x10^9 and 5x10^9 pfus/mL. | |
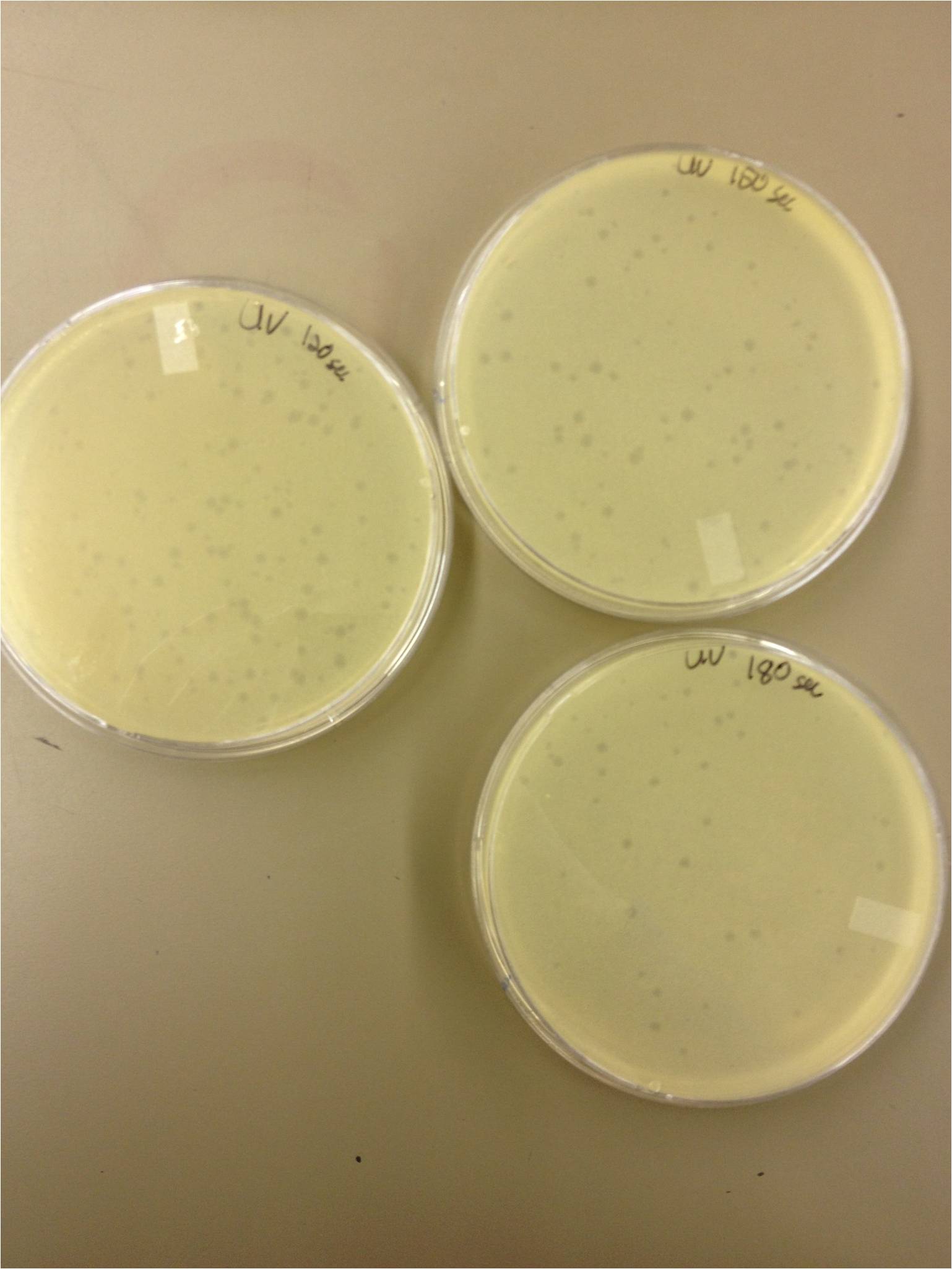
| + | 180 sec = ~50 plaques | ||
| + | 150 sec = ~102 plaques | ||
| + | 120 sec = ~170 plaques | ||
| + | 90 sec = ~2805/6/13- KS, BDM | ||
| + | Today we did a phage titer of 0,-3,-6,-7,-8,-9,-10,-11 We started by putting 1 ul of phage into 1ml of bacteria, we went down from there. We infected each test tube with 10 uL from each dilution level. | ||
| - | + | We also did a UV pre-test of sorts. We put 20ul of phage onto parafilm and exposed it to UV light in the hood of Dr. Breakwell’s lab. At 30 second intervals we took the 20 ul’s off and put it in a half ml of e.coli W3110. We tested every 30 seconds up to 3 minutes. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | We also streaked out the W3110 bacteria from freezer stock onto an LB plate so we can use it to start future bacterial cultures. | |
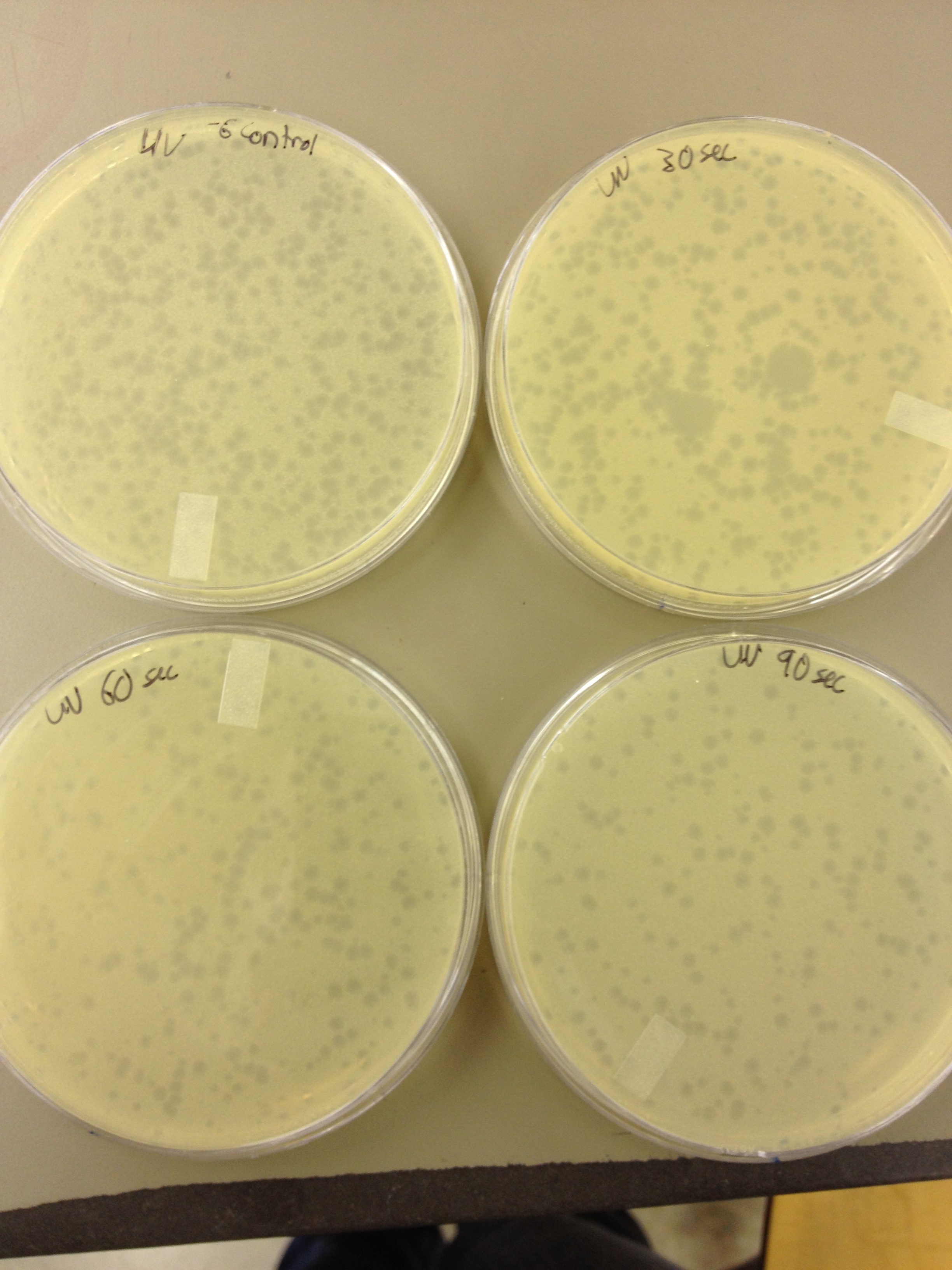
| - | + | plaques | |
| + | 60 sec = ~300 plaques | ||
| + | 30 sec = | ||
| + | 0 sec = ~300-500 plaques | ||
| - | + | Goals soon: | |
| + | Refine liquid culture technique - read online procedure | ||
| + | Pick plaques - what titer do we get on plates from picked plaques? | ||
| + | Make top agar (1.5x to dilute to 0.75x) | ||
| + | Pour LB plates | ||
| + | Purify small plaques from UV test | ||
<br> | <br> | ||
| - | <font | + | <font size="4"> '''5/10/13''' </font> |
| - | + | - Phage similar to T1: | |
| - | + | Rtp | |
| - | + | 46219 | |
| - | + | NC_007603 | |
| - | + | E. coli | |
| + | JBACT 188:1419 | ||
| + | Rogue1 | ||
| + | 45805 | ||
| + | JQ182736 | ||
| + | E. coli | ||
| + | VirolJ 9:2-7 | ||
| + | We found that these two phage have homologous genes to T1, but the nucleotide sequences are not significantly similar. We looked into more T1 articles to try to find someone we can ask to send us a sample, and found this article from 2004 when it was sequenced (http://www.sciencedirect.com/science/article/pii/S0042682203007153). We checked at picultures.com before placing the email, and found that they have both T1 and E. coli B that we can order. We will do this so we can use T1 in our mutagenesis procedure. | ||
<br> | <br> | ||
| - | + | </font> | |
| - | + | ||
| - | + | ||
|} | |} | ||
{{TeamBYUProvoFooter}} | {{TeamBYUProvoFooter}} | ||
Latest revision as of 17:35, 27 September 2013
| Large Phage May - June Notebook: May 1 - May 12 Daily Log
| ||
|
|
5/1/13 Today we scheduled out the rest of spring semester! See Below! Results of looking for recipes M9+ media recipe (grams per liter) KH,PO4 (3.0 g), Na,HPO4 (7.0 g), NaCl (0.5 g), NH4Cl (1.0 g), MgSO4.7HOH (0.246 g), CaCl, (0.011 g), ferric citrate trihydrate (0.6 mg), glucose (4.0 g), and Casamino Acids (10 g; Difco). Phage stock preparation http://myxo.css.msu.edu/ecoli/phagepharm.html
M9 minimal salts media 500g - (or wherever else is cheaper) MgSO4 CaCl ferric citrate trihydrate - not sure if this is necessary...? we only need 0.6 mg per liter and we can't find where it's sold so we'll probably omit it glucose Casamino Acids (500 g) Adenine (we only need 500 mg or so) Uracil (we only need 500 mg or so) Magnesium chloride Chloroform
5/2/13 Today we created a plan for the next little while: May 7-8 and 13-14 Prepare and mutagenize T4 phage Day 1 Start overnight growth of W3110 (http://biology.clc.uc.edu/fankhauser/labs/microbiology/Phage/Phage_Prep.htm) Day 2 before class: Grow E. coli W3110 in H-broth --find substitute-- (supplemented with 20 ug of adenine per ml) to a density of 3 x 10^8 cells/ml - How do we know it's at this density? Day 2 during class: Chill and pellet bacteria Resuspend in at concentration of 1.5 x 10^8 cells/ml in M9+ medium with 5-bromodeoxyuridine (mutagen) and other supplements Add 2 x 10^9 phage particles to 30 mL of bacteria and aerate at 37C until cleared (a few hours). Then add Chloroform. - Good stopping point here May 15 Titer mutagenized phage stocks May 17, 20 Liquid culture of giant phage Day 1 - Start overnight growth of W3110 in M9+ Day 2 - Start mid-log to to density of 4e8 cells/mL. Add mutagenized phages to each culture (MOI of 0.5) - wait 7 minutes - Add ten times the amount of T1 (compared to T4 mutagenized phage) to lyse uninfected cells (preventing giant phage recombination and loss). - T1 has short latent period - Wait seven minutes Find a way to inhibit lysis... Pellet bacteria, resuspend in 25 mL, lyse with Chloroform, treat with DNase and Mg+ Pellet debris May 21 Start centrifugation May 22 Work with small plaques Mutagenize more phage May 24 Titer centrifugation fractions May 29 – UV and plate large phage fractions May 30 Purify large phage further First week of June – take EMs of putative giant phage Second week of June – Plate out lots of samples, look for small plaques, try to purify large phage
5/3/13 -Today we archived our phage stock of T4 Do. Dr. Grose said she couldn’t find T1 easily available so now we are going to look into alternative e.coli phage that has a short latent period.
5/6/13 - Today we did a phage titer of 0,-3,-6,-7,-8,-9,-10,-11 We started by putting 1 ul of phage into 1ml of bacteria, we went down from there. We infected each test tube with 10 uL from each dilution level. We also did a UV pre-test of sorts. We put 20ul of phage onto parafilm and exposed it to UV light in the hood of Dr. Breakwell’s lab. At 30 second intervals we took the 20 ul’s off and put it in a half ml of e.coli W3110. We tested every 30 seconds up to 3 minutes. We also streaked out the W3110 bacteria from freezer stock onto an LB plate so we can use it to start future bacterial cultures.
5/8/13 -We looked at the results for the dilution series (titer) we did on the phage stock we made from the liquid culture and found that there were 5 plaques on the 10^-8 plate. We calculated pfus/mL by doing ( # of plaques ) / (dilution level x mLs infected with). Our stock is between 3x10^9 and 5x10^9 pfus/mL. 180 sec = ~50 plaques 150 sec = ~102 plaques 120 sec = ~170 plaques 90 sec = ~2805/6/13- KS, BDM Today we did a phage titer of 0,-3,-6,-7,-8,-9,-10,-11 We started by putting 1 ul of phage into 1ml of bacteria, we went down from there. We infected each test tube with 10 uL from each dilution level. We also did a UV pre-test of sorts. We put 20ul of phage onto parafilm and exposed it to UV light in the hood of Dr. Breakwell’s lab. At 30 second intervals we took the 20 ul’s off and put it in a half ml of e.coli W3110. We tested every 30 seconds up to 3 minutes. We also streaked out the W3110 bacteria from freezer stock onto an LB plate so we can use it to start future bacterial cultures. plaques 60 sec = ~300 plaques 30 sec = 0 sec = ~300-500 plaques Goals soon: Refine liquid culture technique - read online procedure Pick plaques - what titer do we get on plates from picked plaques? Make top agar (1.5x to dilute to 0.75x) Pour LB plates Purify small plaques from UV test
5/10/13 - Phage similar to T1: Rtp 46219 NC_007603 E. coli JBACT 188:1419 Rogue1 45805 JQ182736 E. coli VirolJ 9:2-7 We found that these two phage have homologous genes to T1, but the nucleotide sequences are not significantly similar. We looked into more T1 articles to try to find someone we can ask to send us a sample, and found this article from 2004 when it was sequenced (http://www.sciencedirect.com/science/article/pii/S0042682203007153). We checked at picultures.com before placing the email, and found that they have both T1 and E. coli B that we can order. We will do this so we can use T1 in our mutagenesis procedure.
| |
 "
"