Exeter/24 July 2013
From 2013.igem.org
Fentwistle (Talk | contribs) (→In gel) |
|||
| (3 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | ==Results of liquid cultures== | + | {{:Team:Exeter/Template/Header}} |
| + | <div class="container"> | ||
| + | <div class="row"> | ||
| + | <div class="span" style="text-align:justify"> | ||
| + | |||
| + | ==Results of liquid cultures== __NOTOC__ | ||
Some of our liquid cultures didn't work; the cells died in the antibiotic. Mildly frustrating. The failed cultures were: | Some of our liquid cultures didn't work; the cells died in the antibiotic. Mildly frustrating. The failed cultures were: | ||
| + | |||
*OmpR (BBa_K098011) | *OmpR (BBa_K098011) | ||
| + | |||
*Lambda inverter system (BBa_Q04510) | *Lambda inverter system (BBa_Q04510) | ||
| Line 47: | Line 54: | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| - | ! Part !! Concentration (ng/ | + | ! Part !! Concentration (ng/µl) |
|- | |- | ||
| CcaR ||324.6 | | CcaR ||324.6 | ||
| Line 71: | Line 78: | ||
| YF1 || 27.1 | | YF1 || 27.1 | ||
|- | |- | ||
| - | | | + | | ''ompC'' || 28.2 |
|} | |} | ||
| Line 79: | Line 86: | ||
==Digestion== | ==Digestion== | ||
| - | We then cut the genes out of the plasmid using Xbal and Pstl. Prepare to run on a gel. | + | We then cut the genes out of the plasmid using <i>Xbal</i> and <i>Pstl</i>. Prepare to run on a gel. |
Each eppendorf contains: | Each eppendorf contains: | ||
| - | * | + | * 12 µl purite water |
| - | * | + | * 2 µl 10X FastDigest Buffer w.Green |
| - | * 0. | + | * 0.5 µl Xbal |
| - | * 0. | + | * 0.5 µl Pstl |
| - | * | + | * 5 µl DNA |
== In gel == | == In gel == | ||
| Line 115: | Line 122: | ||
| 8 || Fix J | | 8 || Fix J | ||
|- | |- | ||
| - | | 9 || OmpR promoter (a.k.a | + | | 9 || OmpR promoter (a.k.a ''ompC'') |
|- | |- | ||
| 10 || Promoter, RBS, #3 | | 10 || Promoter, RBS, #3 | ||
| Line 132: | Line 139: | ||
The gel shows that some of our digestions appear to have worked (our yellow pigment in lane 6 has a clear band at about ~700bp, where would expect) but others appear not to have worked, or have bands that don't match our expected genes. | The gel shows that some of our digestions appear to have worked (our yellow pigment in lane 6 has a clear band at about ~700bp, where would expect) but others appear not to have worked, or have bands that don't match our expected genes. | ||
| - | We | + | ==Cph8 gel== |
| + | |||
| + | We're still uncertain about the presence of a <i>PstI</i> cut site in BBa_K322124 (cph8, our red light sensor), so we want to try cutting it with different combination of <i>EcoRI</i>, <i>XbaI</i>, <i>SpeI</i> and <i>PstI</i>. We have five replicates of cph8 plasmid from previous MiniPreps. | ||
* cph8 #3 | * cph8 #3 | ||
| Line 144: | Line 153: | ||
* cph8 #7 | * cph8 #7 | ||
| - | + | Unfortunately, they were only cut with <i>Xbal</i> and <i>Pstl</i> (Fran...), but this gel can be correctly run tomorrow with relative ease. | |
| + | |||
| + | The lanes were run in the order above, flanked by a 1kb Gene Ruler. | ||
| + | |||
| + | Take me back to the [https://2013.igem.org/Team:Exeter/Notebook notebook]. | ||
| - | + | </div> | |
| + | </div> | ||
| + | {{:Team:Exeter/Template/Footer}} | ||
Latest revision as of 20:24, 2 October 2013
Results of liquid cultures
Some of our liquid cultures didn't work; the cells died in the antibiotic. Mildly frustrating. The failed cultures were:
- OmpR (BBa_K098011)
- Lambda inverter system (BBa_Q04510)
Minipreps using the Qiagen kit
We miniprepped:
- CcaR
- Promoter, RBS x 3 replicates
- CcaS
- B0034
- Magenta
- OmpR promoter
- OmpR
- YF1
- Lambda inverta
- Fix J
- Yellow
- Fix J promoter
Glycerol stocks
We made glycerol stocks of what is above, then stored them at -80oC
For future reference, to get glycerol stocks back as plates, take a loop and streak on gel. Has to be taken from frozen glycerol stocks, which are immediately returned to the freezer.
We then made a gel for digests using ethidium bromide.
NanoDrop from miniprep
| Part | Concentration (ng/µl) |
|---|---|
| CcaR | 324.6 |
| CcaS | 264.4 |
| Yellow | 64.3 |
| Magenta | 102.7 |
| RBS | 48.9 |
| Promoter, RBS, #1 | 21.5 |
| Promoter, RBS, #2 | 22.6 |
| Promoter, RBS, #3 | 22.0 |
| Fix L | 31.9 |
| Fix J | 35.4 |
| YF1 | 27.1 |
| ompC | 28.2 |
No SureClean was needed.
Digestion
We then cut the genes out of the plasmid using Xbal and Pstl. Prepare to run on a gel.
Each eppendorf contains:
- 12 µl purite water
- 2 µl 10X FastDigest Buffer w.Green
- 0.5 µl Xbal
- 0.5 µl Pstl
- 5 µl DNA
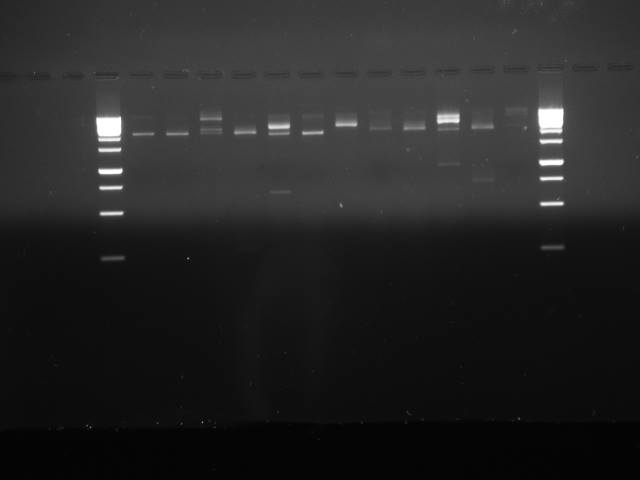
In gel
| Lane | Content |
|---|---|
| 1 | Ladder (1kb Gene Ruler) |
| 2 | Promoter, RBS, #1 |
| 3 | B0034 |
| 4 | CcaS |
| 5 | Fix J promoter (a.k.a Fix L) |
| 6 | Yellow |
| 7 | Promoter, RBS, #2 |
| 8 | Fix J |
| 9 | OmpR promoter (a.k.a ompC) |
| 10 | Promoter, RBS, #3 |
| 11 | Magenta |
| 12 | CcaR |
| 13 | YF1 |
| 14 | Ladder (1kb Gene Ruler) |
The gel shows that some of our digestions appear to have worked (our yellow pigment in lane 6 has a clear band at about ~700bp, where would expect) but others appear not to have worked, or have bands that don't match our expected genes.
Cph8 gel
We're still uncertain about the presence of a PstI cut site in BBa_K322124 (cph8, our red light sensor), so we want to try cutting it with different combination of EcoRI, XbaI, SpeI and PstI. We have five replicates of cph8 plasmid from previous MiniPreps.
- cph8 #3
- cph8 #4
- cph8 #5
- cph8 #6
- cph8 #7
Unfortunately, they were only cut with Xbal and Pstl (Fran...), but this gel can be correctly run tomorrow with relative ease.
The lanes were run in the order above, flanked by a 1kb Gene Ruler.
Take me back to the notebook.
 "
"