Team:Heidelberg/Delftibactin/DelH
From 2013.igem.org
Nils.kurzawa (Talk | contribs) m |
|||
| Line 68: | Line 68: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 2</h1> | <h1>Week 2</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Using the designed primers DelH_f1_PacI_fw and DelH_f1_SalI_rev, the first PCRs to amplify DelH F1 as well as DelH_f2_SalI_fw and DelH_f2_KpnI_rev to amplify DelH F2 were performed and conditions optimized. Additionally, necessary backbone fragments pSB6A1 and lacZ were restriction digested. New BioBricks to obtain the AraC promotor (I13453 and K206000) were chosen. |
| - | + | ||
| - | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 79: | Line 77: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 3</h1> | <h1>Week 3</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Since the reverse backbone primer DN08:AraCbb_PacI_rev did not work, a new one (DN06:AraCbb_PacI_rev2) was ordered, together with the colony PCR screening primer DN07:Screen_DelH_rev and DN13:Screen_DelH_fw to check for correct ligation of the DelH fragment F1. |
| - | + | ||
| - | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 90: | Line 86: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 4</h1> | <h1>Week 4</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">The elongation of the pSB6A1-AraC-lacZ backbone urned out to be troublesome. So in week 4, we performed different restriction digests with our final backbone pSB6A1-AraC-lacZ as well as with the former construct pSB1C3-AraC-lacZ to ensure identity of the backbone. Besides, the amplification of the DelH fragment F1 was planned: It is amplified divided in 2 subfragments - fragment F1a and fragment F1b. Each one of the fragments has the size of 5 Kb. </p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 99: | Line 94: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 5</h1> | <h1>Week 5</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In this week, we started all over again to assemble the backbone pSB6A1-AraC-lacZ: digesting AraC, lacZ and PSB6A1 based on the precedently amplified fragments. The amplification of DelH was continued and we succesfully amplified all 3 fragments and gel extracted them. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 108: | Line 103: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 6</h1> | <h1>Week 6</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Because we amplified all DelH-fragments needed and assembled the backbone in week 5, this week, we assembled the final plasmid pHM01. Therefore, every fragment was digested with two distinct enzymes, and then ligated. The ligated plasmid pHM01 was purified and electroporated in two separate DH10ß aliquots. Unfortunately, the screening via colony-PCR was negative, so none of the transformed E.coli successfully received the plasmid. </p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 117: | Line 111: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 7</h1> | <h1>Week 7</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In week 7, we reamplified the DelH fragments used for the transformation in week 6. We found out that amplification of fragments F1a and F1b was not reproducible. As conclusion, we designed new primers for DelH, which are capable to amplify the beginning of DelH in a more efficient and specific manner. The overview summarizes the primers we ordered and their performance in amplificating DelH F1a. Primer DN11 worked really well and is used in the next experiments. </p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 127: | Line 120: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 8</h1> | <h1>Week 8</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After improving the amplification of the beginning of DelH (fragment 1a with the primer DN11), we used a higher concentration in the ligation assembling the pHM01 plasmid. The transformation of DH10ß cells was performed with electroporation. Four colonies were positive in the screening-PCR, but did not express lacZ (no blue color). To quantify if DelH is possibly expressed anyhow, we plan a SDS-PAGE and induce the bacteria with higher concentrations of arabinose and X-Gal. |
| - | + | </p> | |
</div> | </div> | ||
</div> | </div> | ||
| Line 137: | Line 130: | ||
<h1>Week 9</h1> | <h1>Week 9</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">The SDS-PAGE showed no clear band at ~600 kDa, so we conclude that there is most probably no DelH expression in the analyzed colonies. To confirm the result, a restriction digest was performed. The colonies did not show the expected pattern, so they indeed did not contain the desired plasmid. To perform a new assembly of the plasmid pHM01, the fragments of DelH and the backbone had to be reamplified. The amplifications of the DelH fragments went well, but the backbone caused again some difficulties and it was not possible to amplify it. |
| - | + | ||
</p> | </p> | ||
| Line 148: | Line 140: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 10</h1> | <h1>Week 10</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">When rechecking all amplified fragments, we did not find what we expected. On the one hand, the concentrations of the fragments were too low and could not be seen on the gel. On the other hand, a re-amplification of the fragments with the appropriate primers was not successful. Additionally, a new polymerase (Phusion Flash) was used. Interestingly for the DelH 1b fragment, the fragment amplified from the genomic DNA was slightly larger than the amplified fragments from itself. |
| - | + | ||
| - | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 159: | Line 149: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 11</h1> | <h1>Week 11</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Based on this week's experiments and in silico analysis to validate the fragments DelH F1a, F1b and F2, we discarded the restriction digest and ligation strategy for the assembly of the DelH plasmid. Instead, we chose Gibson assembly as alternative method, developed a strategy and designed primers accordingly. We are positive, to finally assemble pHM02. |
| - | + | ||
| - | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 170: | Line 158: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 12</h1> | <h1>Week 12</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">The primers arrived and we started amplifying the Gibson fragments using different approaches. Since G1 could not be PCR amplified, we decided to divide G1 into sub fragments G1a and G1b, for which we had designed and ordered primers already. The amplification of fragment G2 worked well and was produced in reasonable amounts. Alternatively, we also produced the entire DelH as one fragment G0, as well as further sub fragments DelH G1/2a, G1b/2 and G2b. Amplification of G1b/2a did not work out. Due to the failure of the restriction digest strategy, we further analyzed the backbone pSB6A1-AraC-lacZ, which we decided to discard and instead, use pSB6A1 and BBa_J04450, which is already available in the parts registry. </p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 179: | Line 166: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 13</h1> | <h1>Week 13</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In order to realize the new strategy using the already existing backbone from the parts registry pSB6A1-lacZ-mRFP, we designed two new primers for the backbone amplification and created a map of pHM03. |
| + | The plasmid for the backbone was obtained from the registry, transformed in E.coli and minipreped. The Gibson fragment of the backbone was successfully amplified, together with Gibson fragments DelH G0, G1 and G2b. </p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 188: | Line 176: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 14</h1> | <h1>Week 14</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In week 14, we've tested numerous colonies from last week's Gibson assembly 28-07 using DelH G0 as well as G1/2a and 2b by screening-PCR. None of the analyzed colonies carried a correct plasmid. We therefore performed another Gibson assembly 01-08 and again screened numerous clones. We found few possibly correct ones that will be further analyzed next week. |
| - | + | Additionally, the new D. acidovorans strain SPH1, whose sequence is available in GenBank, was ordered. We will have to amplify all fragments from this strain again as soon as it arrives. </p> | |
</div> | </div> | ||
</div> | </div> | ||
| Line 197: | Line 185: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 15</h1> | <h1>Week 15</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We further tested colonies from last weeks Gibson assemblies using DelH G0 as well as G1/2a and 2b. Unfortunately, they were all negative in the screening-PCR. Therefore, we amplified our Gibson fragments again and performed more Gibson assemblies. Yet again, there were no positive colonies in the colony-PCR. </p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 207: | Line 194: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 16</h1> | <h1>Week 16</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We further characterized the DelH plasmid created using Gibson assembly. Unfortunately, none of the screened colonies was for sure positive. Selection of red colonies was not clear, and PCR screened colonies were all negative. |
| - | + | In order to avoid high background during screening of the colonies, we decided to run two different strategies. For first approach pHM04, we will amplify the backbone without mRFP, avoiding backbone reassembly due to ribosome binding site homology. In the second strategy pHM05, we will additionally introduce a tetracycline resistance, to ensure integration of the insert. | |
| + | In addition to the primers for the new strategies, we designed a new screening primer at the end of DelH. </p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 217: | Line 205: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 17</h1> | <h1>Week 17</h1> | ||
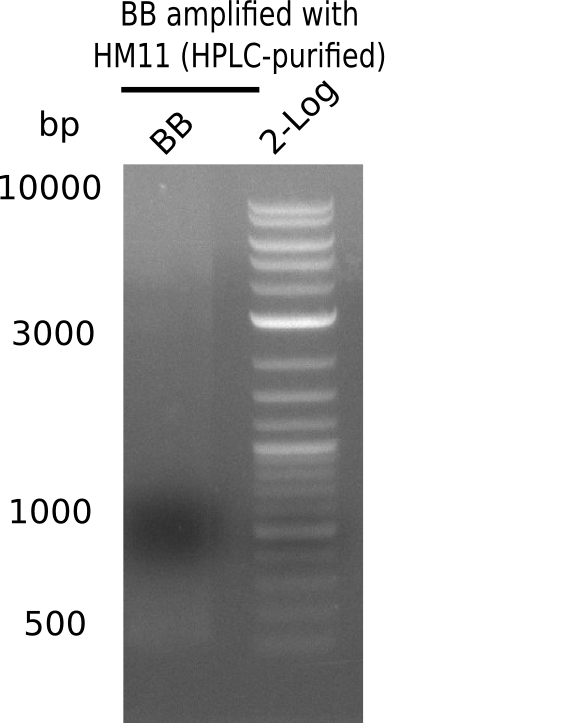
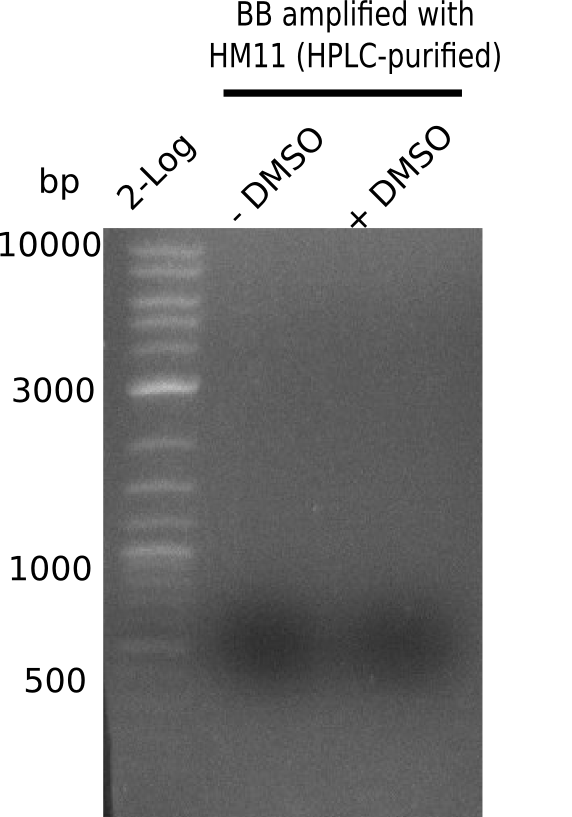
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After performing different Gibson assemblies and screening always resulted in negative clones, the possible explanation could be a religation of the backbone fragment pSB6A1 allowing E.coli to survive and express mRFP. This week, our aim is to design a new construct without mRFP, so that we can exclude the red colonies from the screening. Therefore, we are going to use a new reverse primer for the backbone including only the terminator of the mRFP, but not the mRFP. The primers for the backbone are HM11 & HM17. The construct is named pHM04 and further explained in week 16. </p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 227: | Line 214: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 18</h1> | <h1>Week 18</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Because the constructing pHM04 did not result in any positive clone (see experiments week 17), we will follow the idea to introduce a tetracycline resistance to the ampicillin backbone as additional selection marker for successful assembly. So we have to screen only colonies, which are white (because of the exclusion of mRFP) and grow on plates containing tetracycline. Furthermore, we decided to amplify DelH in various fragments to increase Gibson assembly efficiency. Therefore, we also ordered new screening primers. The primers were designed as shown in the following table. </p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 237: | Line 223: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 19</h1> | <h1>Week 19</h1> | ||
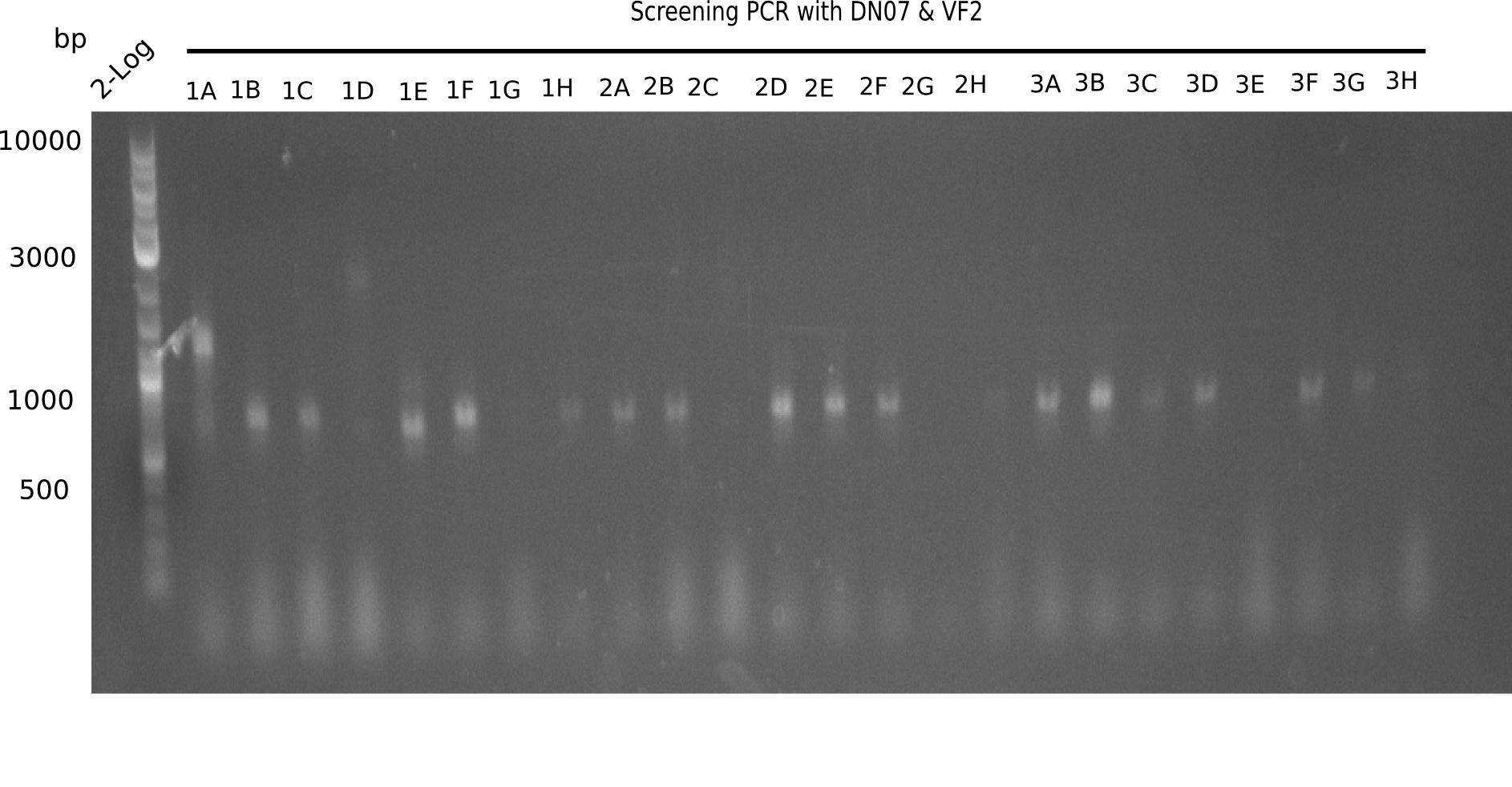
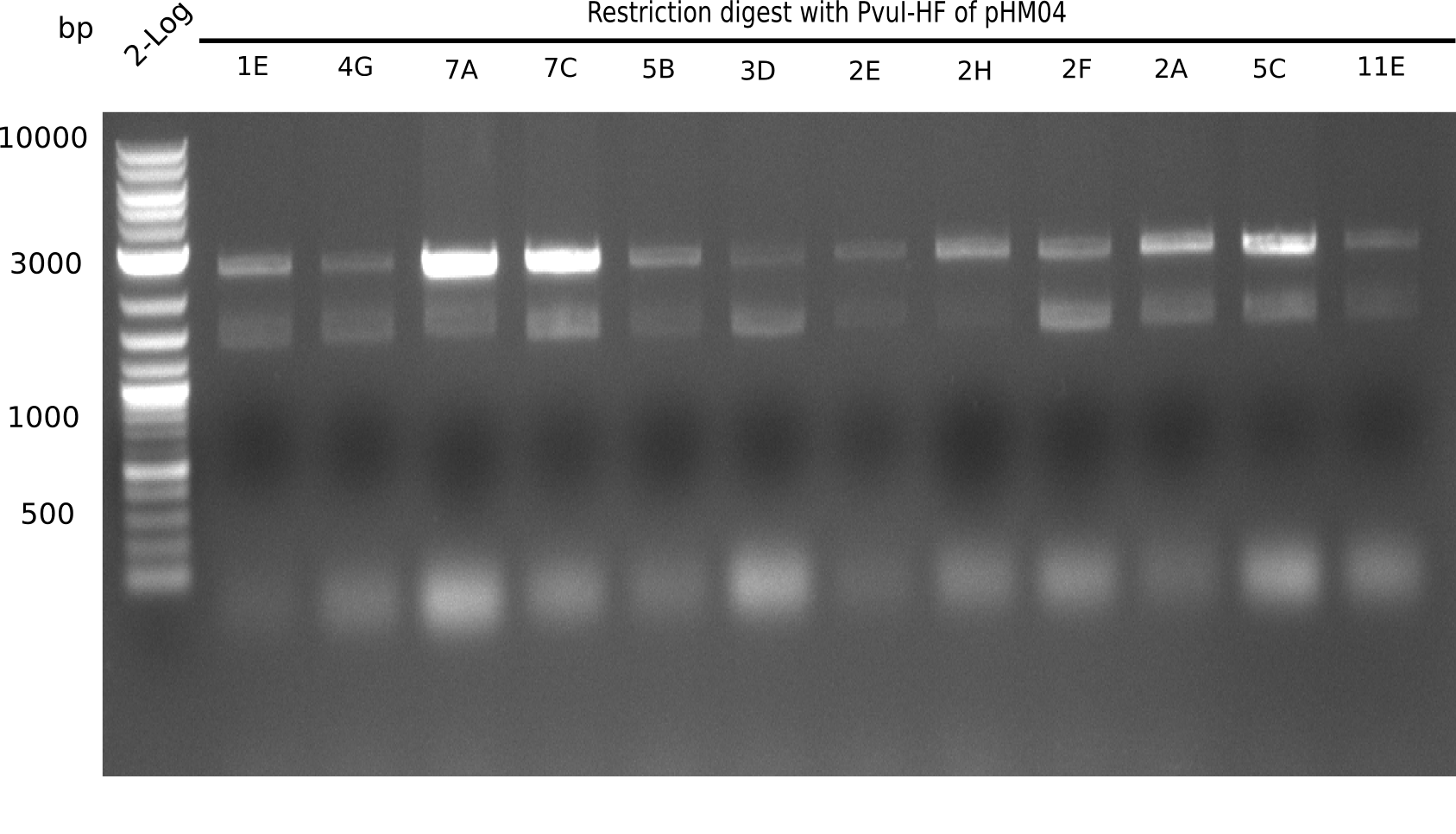
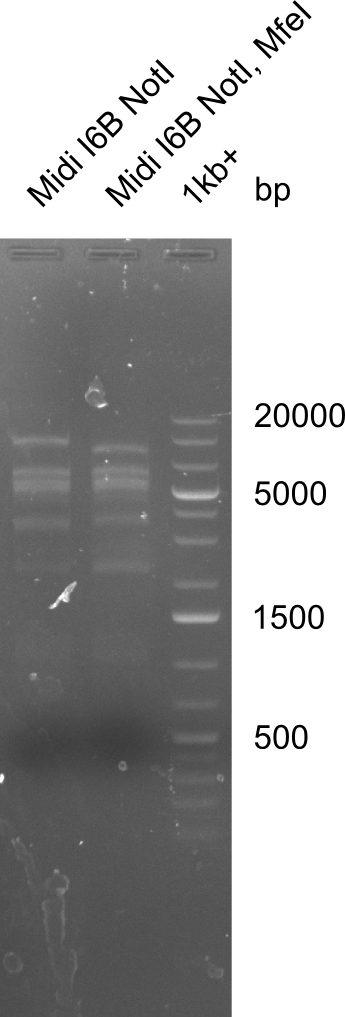
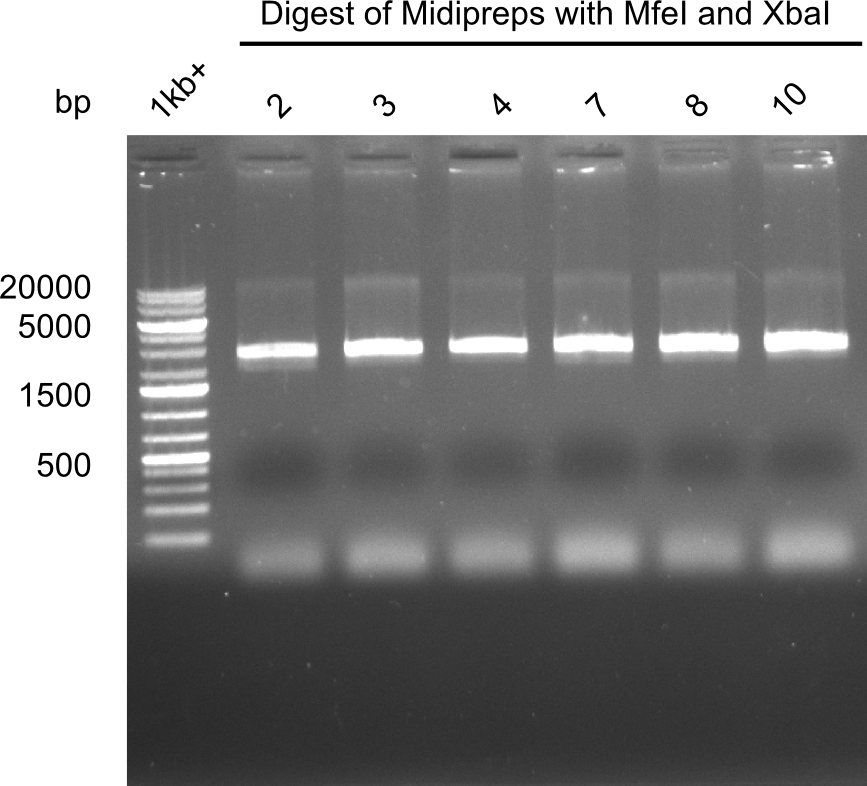
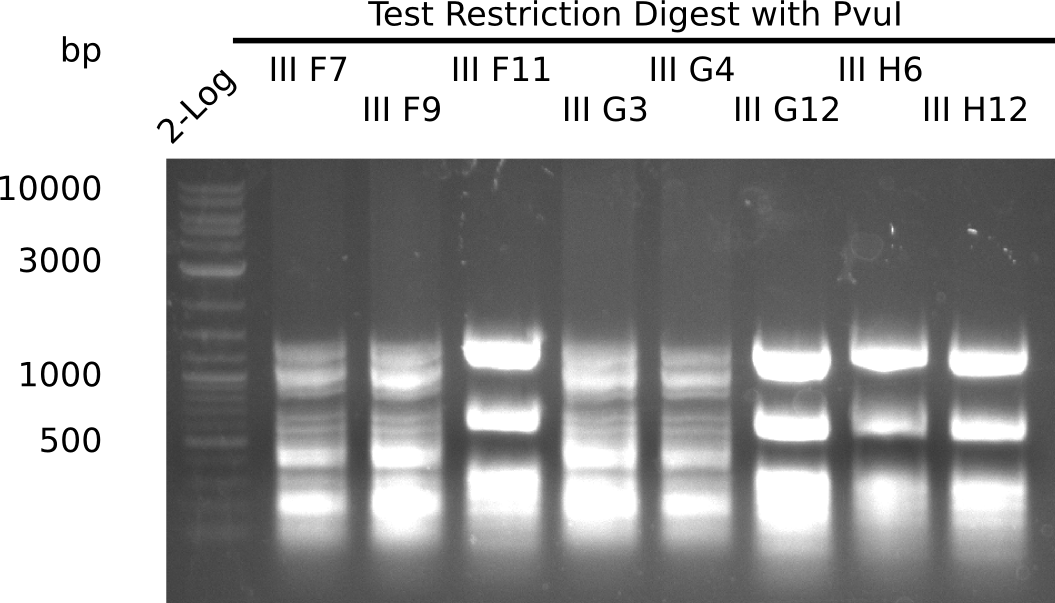
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After a successful electroporation, we screen numerous clones by colony-PCR and test restriction digest (PvuI-HF). Positive clones are send for sequencing. The sequencing tells us if the DelH Gibson assembly actually worked. Therefore, we send the midipreped plasmids with the reverse DN07 primer or VF2, covering the start sequence of DelH and the end of the pSB6A1 backbone without mRFP (pHM04). |
| - | + | The screening-PCR showed different positive colonies. After performing the restriction digest, some colonies were discarded and the rest was sent in for sequencing. We did not find any positive clone without a truncating mutation at the beginning of DelH (in the primer region). </p> | |
</div> | </div> | ||
</div> | </div> | ||
| Line 247: | Line 233: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 20</h1> | <h1>Week 20</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Again, the screening-PCRs resulted in many positive colonies. After the restriction digest, many colonies were kicked out. The remaining minipreped colonies were sent in for sequencing. Yet, we did not identify any positive clone which had no mutation at the beginning of DelH (in the primer region). </p> |
<p><a class="btn btn-large btn-primary" href="#">Browse gallery</a></p> | <p><a class="btn btn-large btn-primary" href="#">Browse gallery</a></p> | ||
</div> | </div> | ||
| Line 257: | Line 243: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 21</h1> | <h1>Week 21</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Frustratingly, none of the analyzed clones showed a correct sequence. Probably, DelH by itself is toxic for E.coli and thus, only aberrant clones survive. We suspect the low quality of primers as reason for high number of mutations. Therefore, we will order HPLC purified primers. |
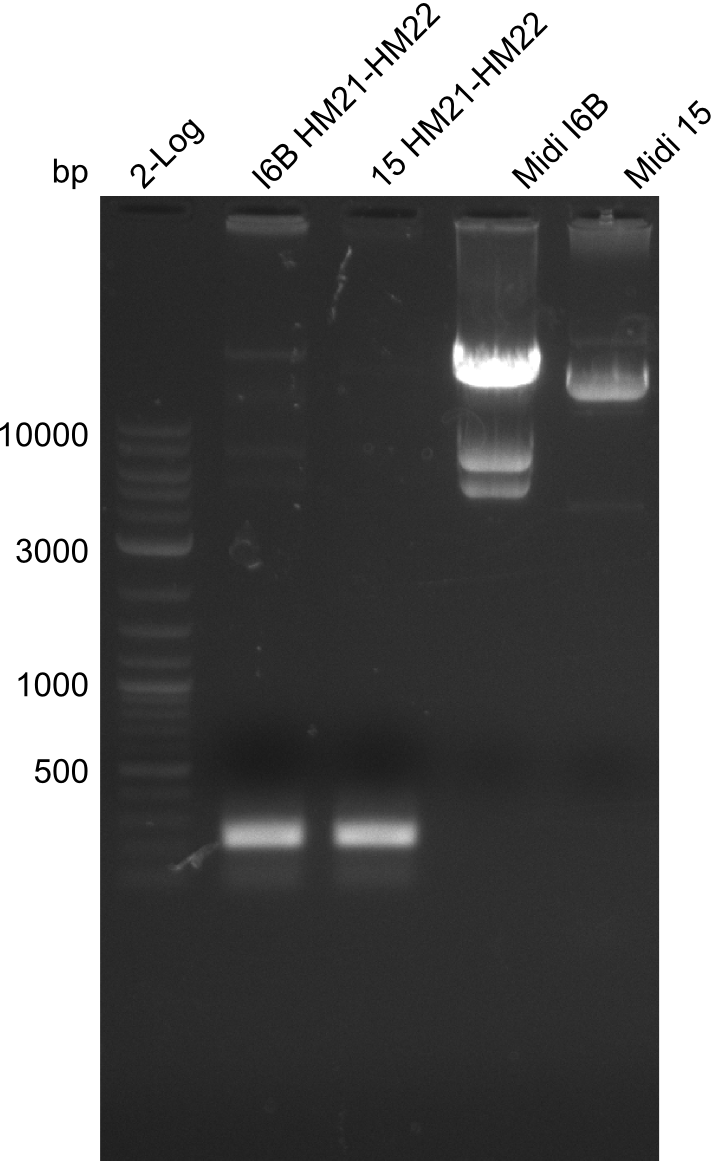
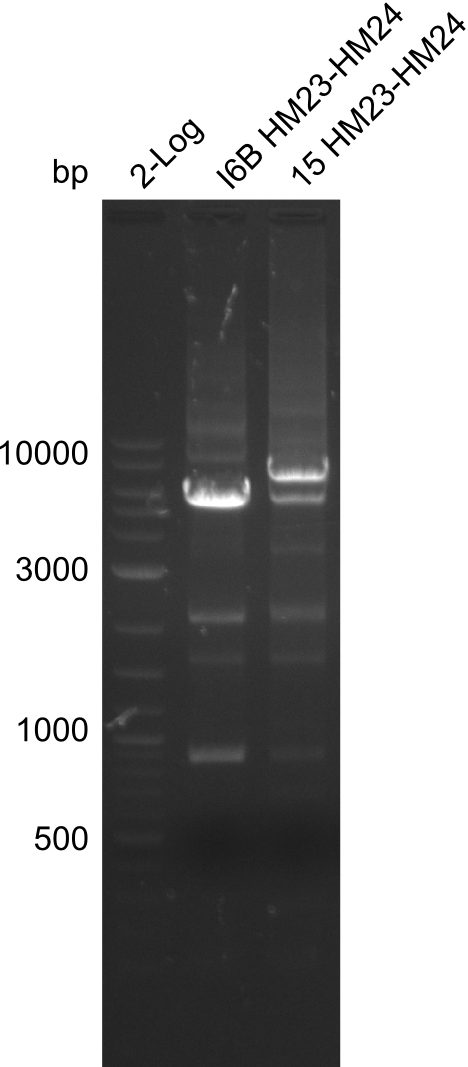
| - | + | Additionally, we tried to eliminate the mutations in DelH clones I 6b and 15 by mutagenesis. Herefore, primers were ordered. </p> | |
</div> | </div> | ||
</div> | </div> | ||
| Line 267: | Line 253: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 22</h1> | <h1>Week 22</h1> | ||
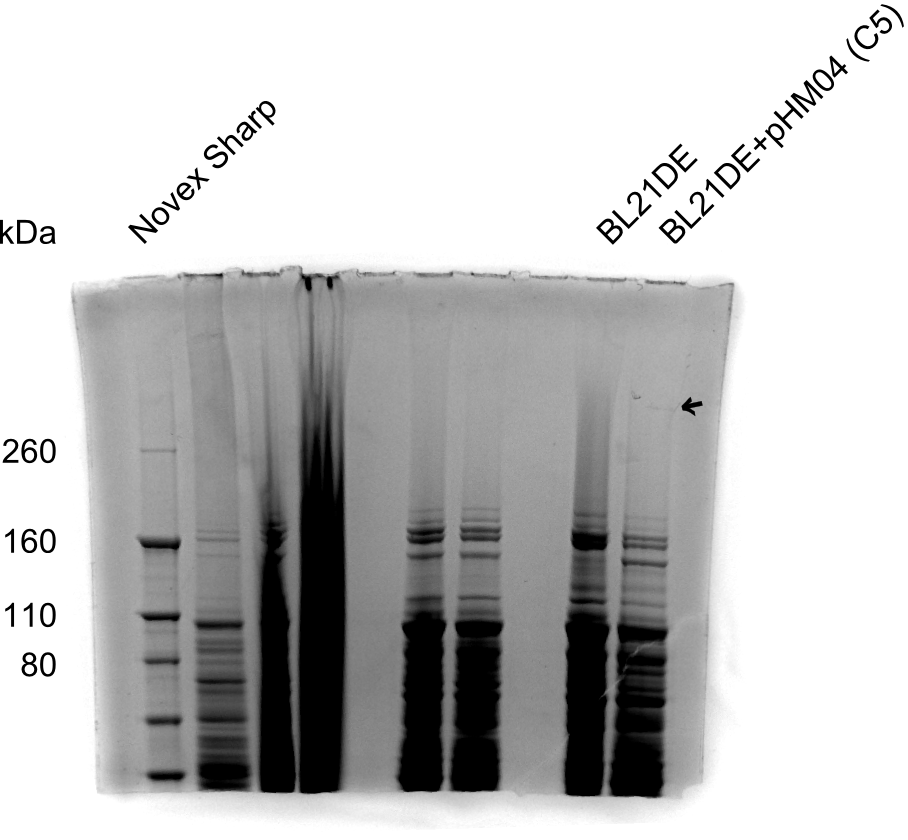
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">So far, we failed to find a single correct DelH clone. We suspect the separate DelH module to be toxic for the E. coli. Therefore, they select for mutated plasmids. In order to reduce the selection pressure, we used E. coli BL21 DE3, known for increased expression of the lac repressor. This strategy also did not result in any correct clone. One reason might be, that the E. coli BL21 DE3 we obtained actually are BL21 DE3 pLys, which are unfortunately Chloramphenicol resistant due to additional plasmid and thus not useful for screening and amplification of DelH construct. |
| - | + | We got new and correct E. coli BL21 DE3 as well as NEB turbo, which significantly overexpress the lac repressor. We however found their lacZ-controlled expression to be very leaky, in contrast to E. coli BL21 DE3. | |
| + | Since the Gibson assembly of DelH using the HPLC purified primers also exclusively resulted in mutated clones, we developed two new cloning strategies to avoid expression of DelH. The first strategy uses a weak promoter and RBS, the second introduces DelH in a ccdB helper construct. Last but not least, we continued working on the mutagenesis approach to eliminate the mutation in clone C5. It was sent for sequencing... </p> | ||
</div> | </div> | ||
</div> | </div> | ||
Revision as of 07:46, 2 October 2013


Del H. This bitchy 18 kbp fragment.
Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet.
Week 1
In order to transfer the gold precipitating NRPS from D. acidovorans to E.coli, the necessary modules will be amplified from the D. acidovorans genome and assembled as plasmids. Due to its large size of 18 Kb, the module DelH will be expressed on a separate plasmid. A strategy was developed, primers designed accordingly and necessary BioBricks obtained from the distribution. The D. acidovorans was obtained from the DSMZ and cultured in Acidovorax complex medium.
Week 2
Using the designed primers DelH_f1_PacI_fw and DelH_f1_SalI_rev, the first PCRs to amplify DelH F1 as well as DelH_f2_SalI_fw and DelH_f2_KpnI_rev to amplify DelH F2 were performed and conditions optimized. Additionally, necessary backbone fragments pSB6A1 and lacZ were restriction digested. New BioBricks to obtain the AraC promotor (I13453 and K206000) were chosen.
Week 3
Since the reverse backbone primer DN08:AraCbb_PacI_rev did not work, a new one (DN06:AraCbb_PacI_rev2) was ordered, together with the colony PCR screening primer DN07:Screen_DelH_rev and DN13:Screen_DelH_fw to check for correct ligation of the DelH fragment F1.
Week 4
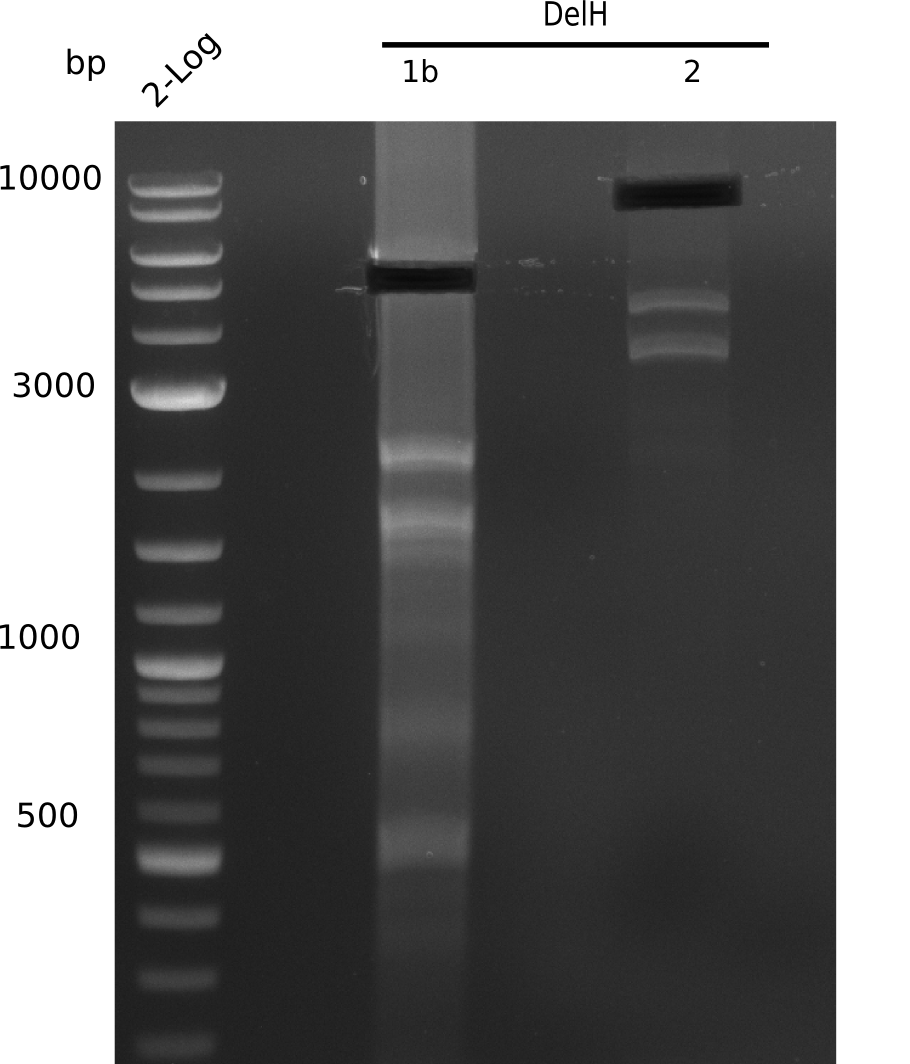
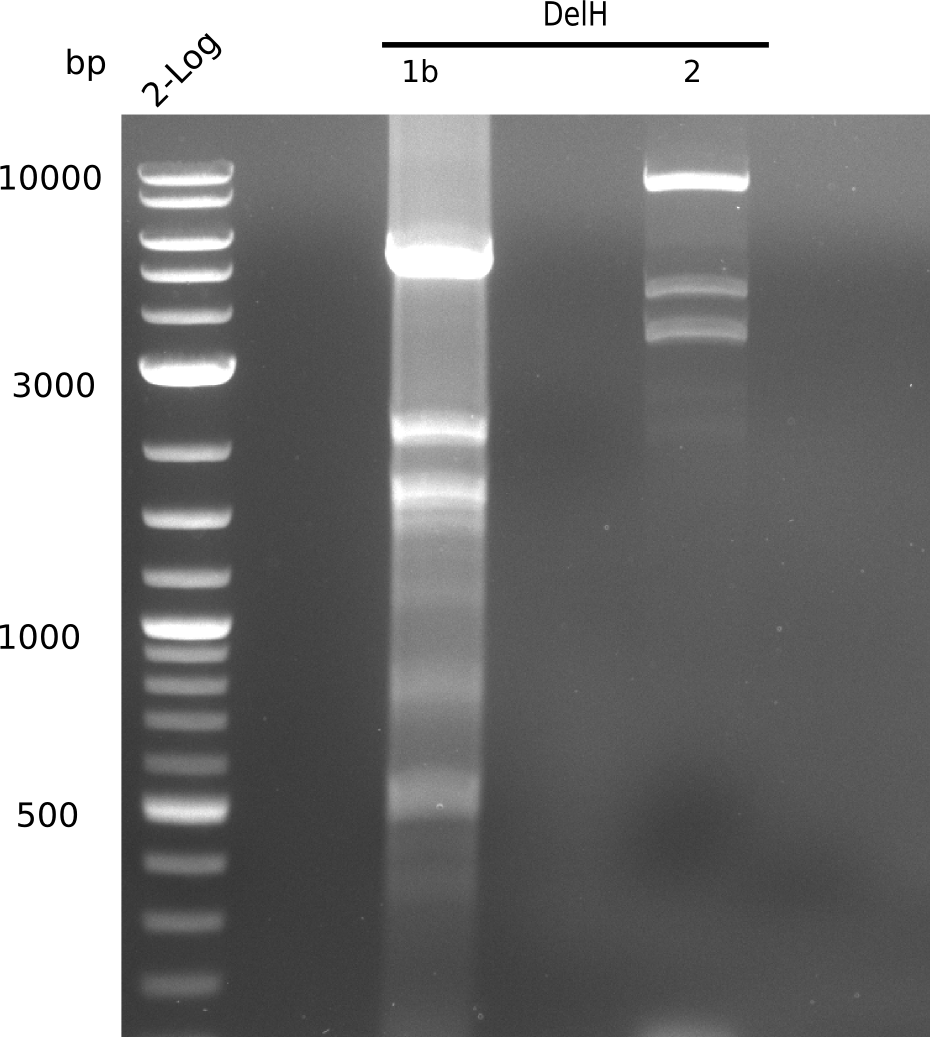
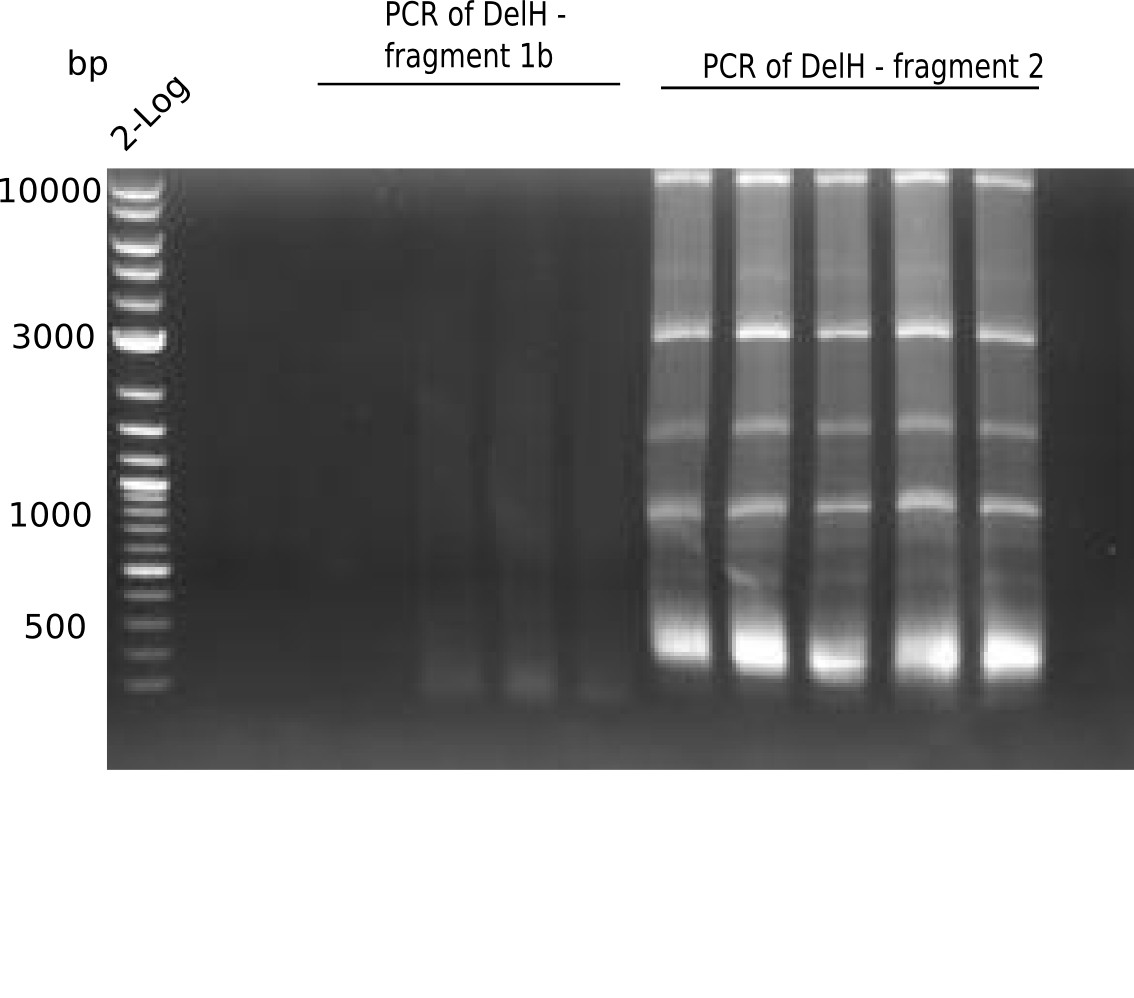
The elongation of the pSB6A1-AraC-lacZ backbone urned out to be troublesome. So in week 4, we performed different restriction digests with our final backbone pSB6A1-AraC-lacZ as well as with the former construct pSB1C3-AraC-lacZ to ensure identity of the backbone. Besides, the amplification of the DelH fragment F1 was planned: It is amplified divided in 2 subfragments - fragment F1a and fragment F1b. Each one of the fragments has the size of 5 Kb.
Week 5
In this week, we started all over again to assemble the backbone pSB6A1-AraC-lacZ: digesting AraC, lacZ and PSB6A1 based on the precedently amplified fragments. The amplification of DelH was continued and we succesfully amplified all 3 fragments and gel extracted them.
Week 6
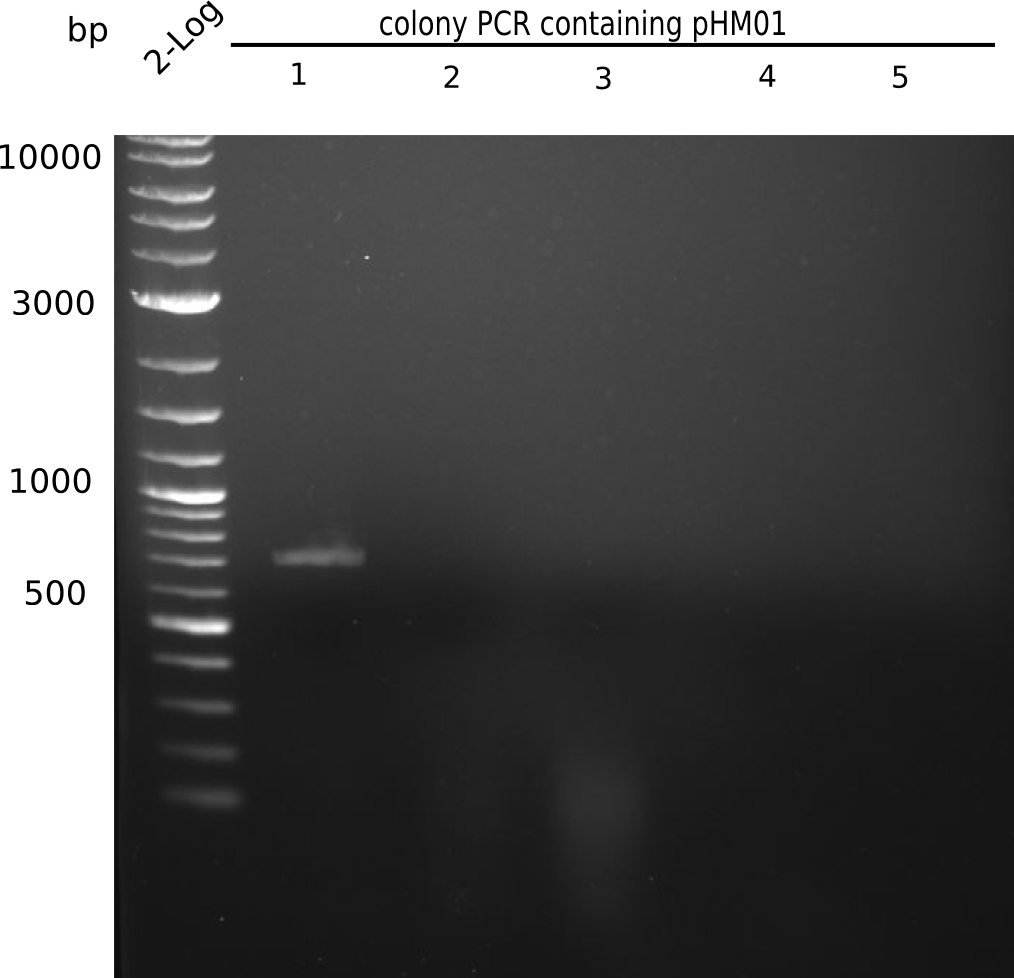
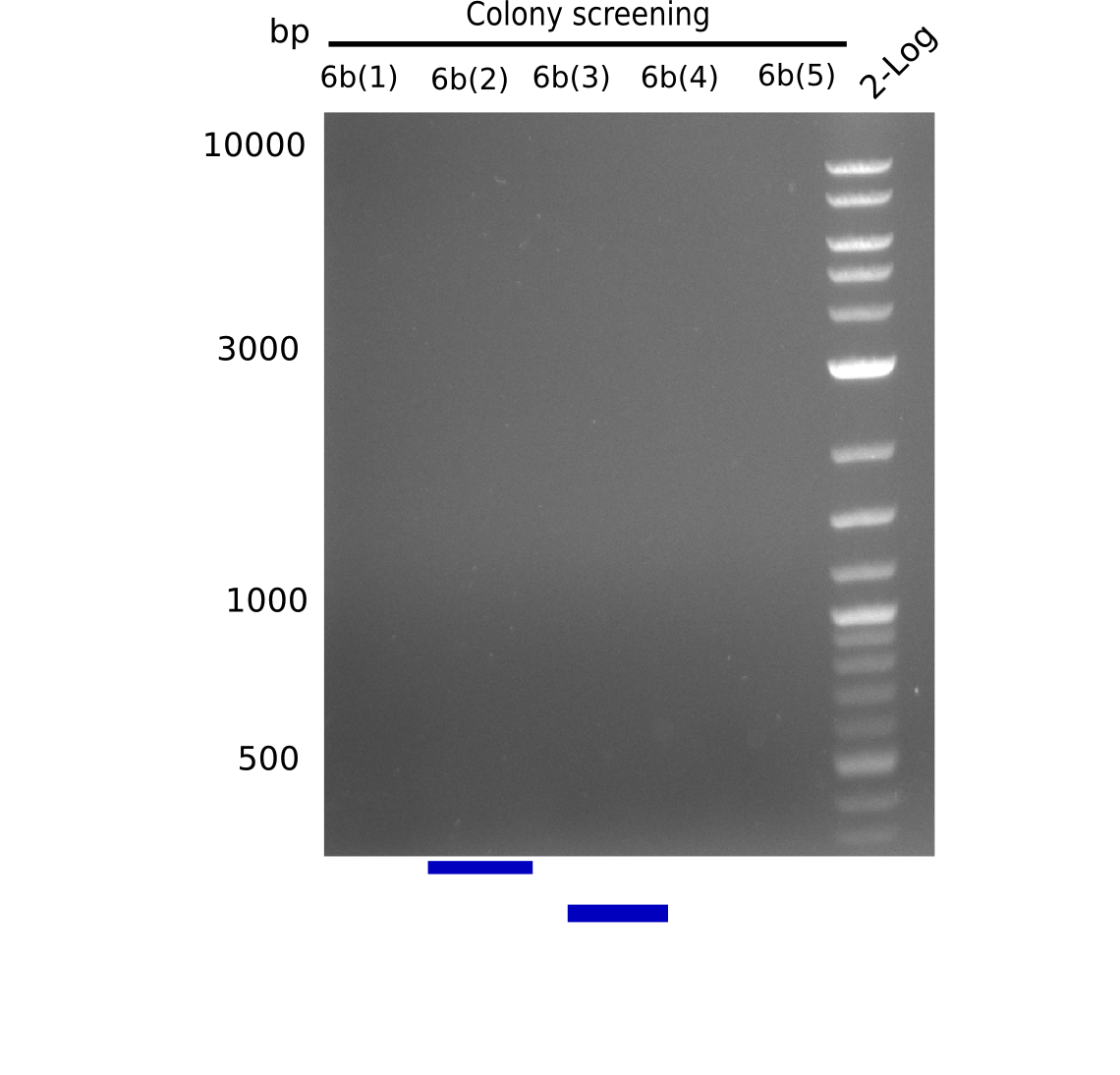
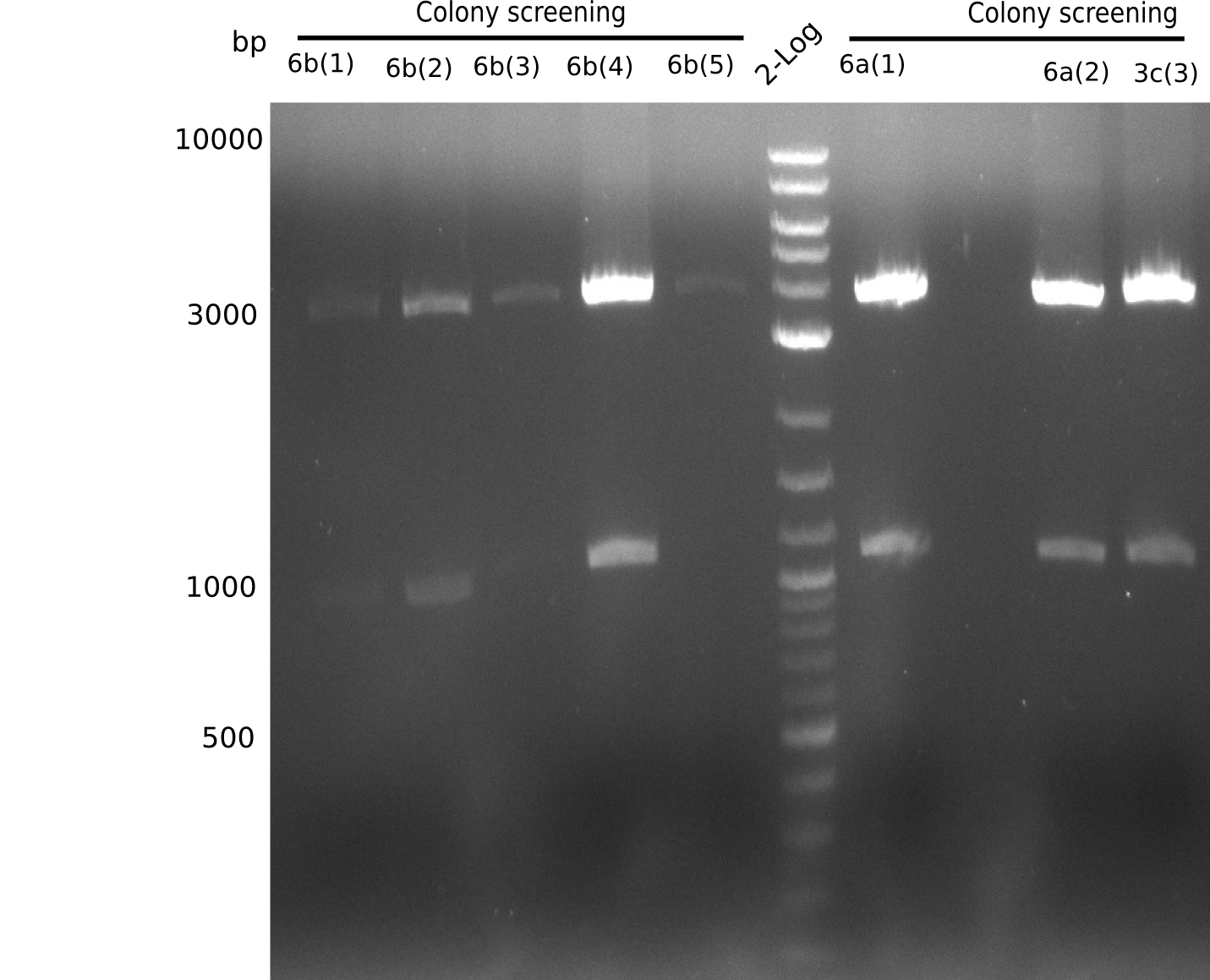
Because we amplified all DelH-fragments needed and assembled the backbone in week 5, this week, we assembled the final plasmid pHM01. Therefore, every fragment was digested with two distinct enzymes, and then ligated. The ligated plasmid pHM01 was purified and electroporated in two separate DH10ß aliquots. Unfortunately, the screening via colony-PCR was negative, so none of the transformed E.coli successfully received the plasmid.
Week 7
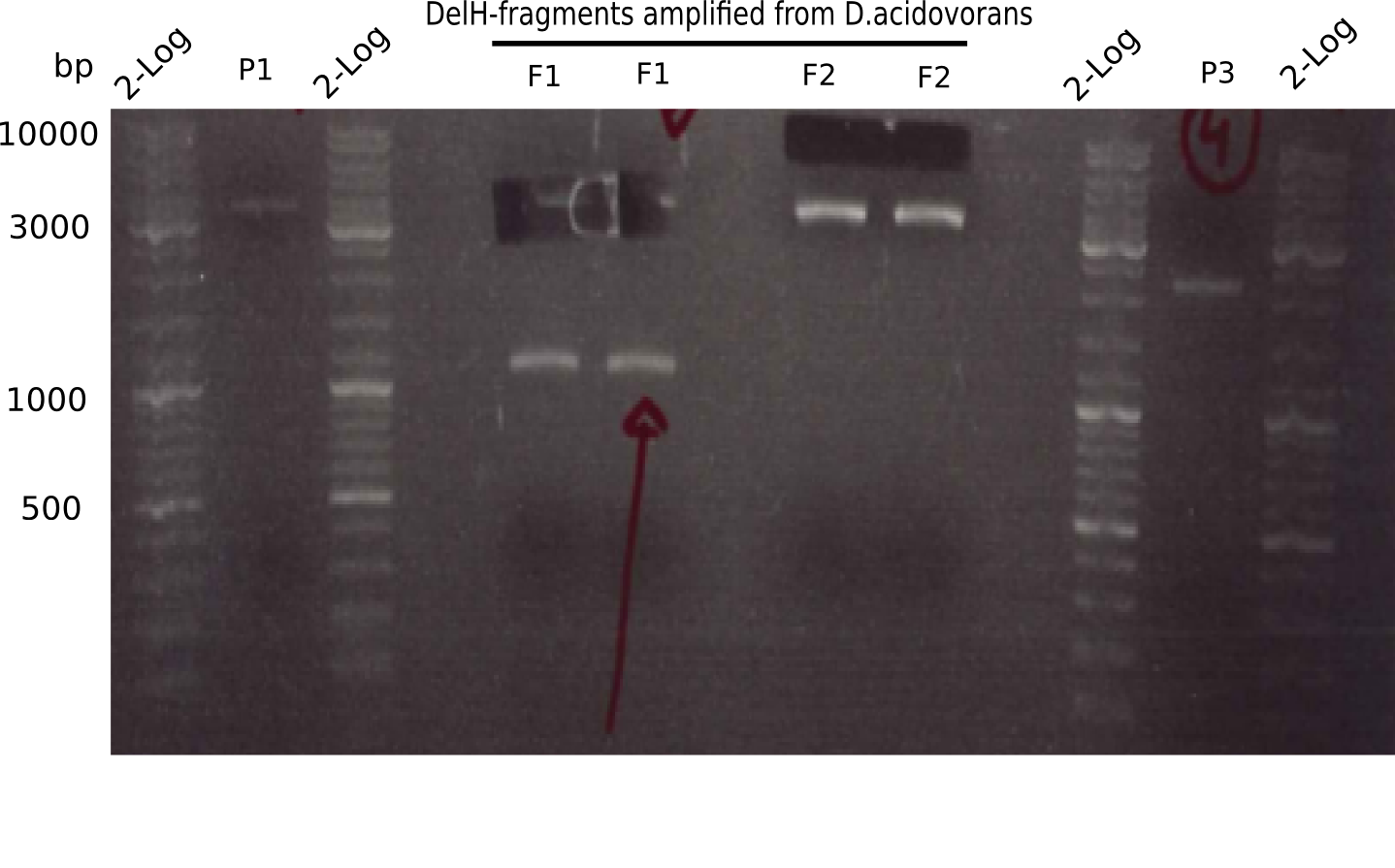
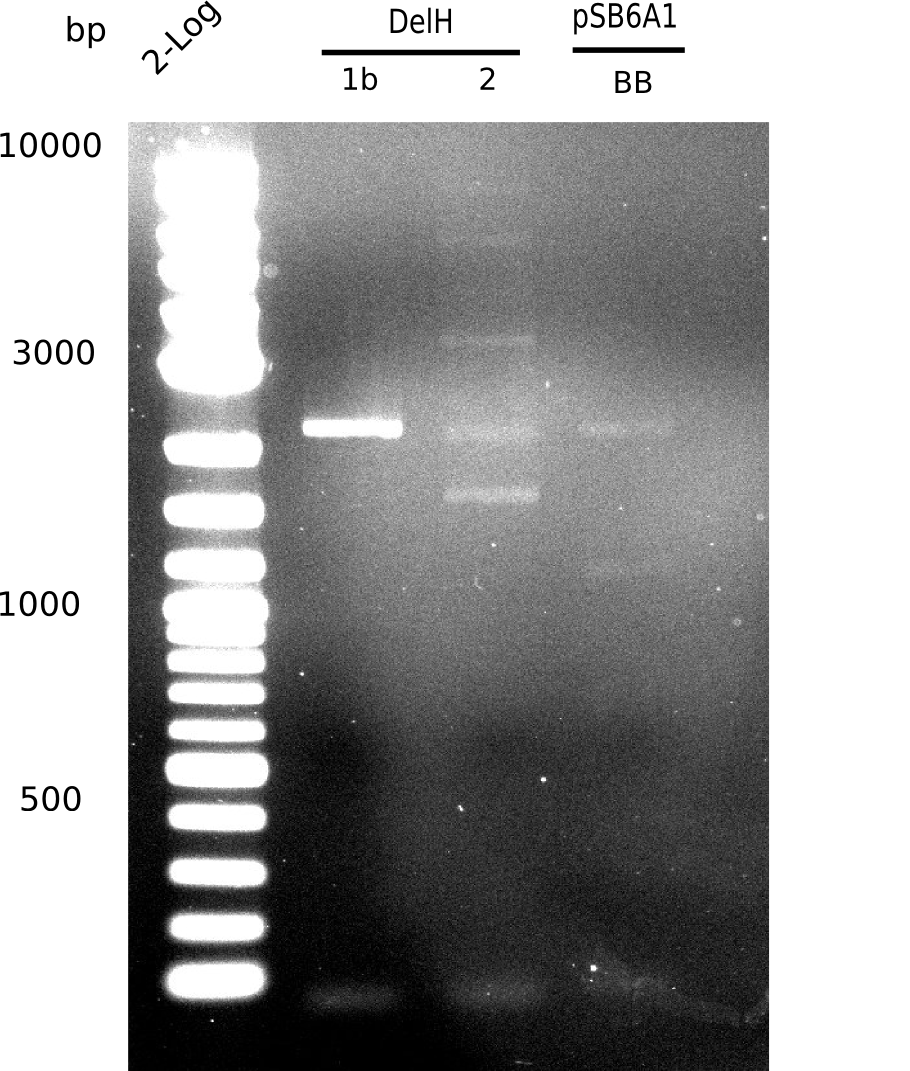
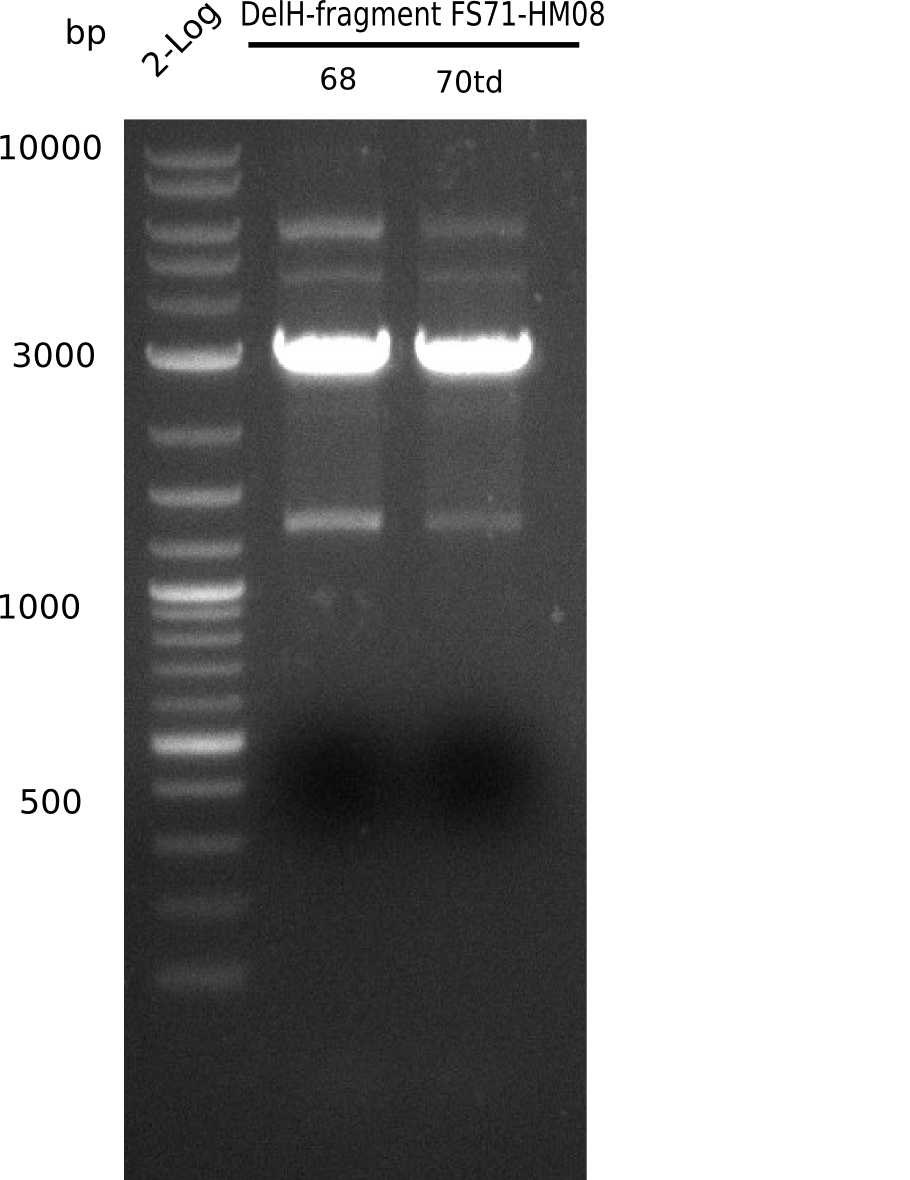
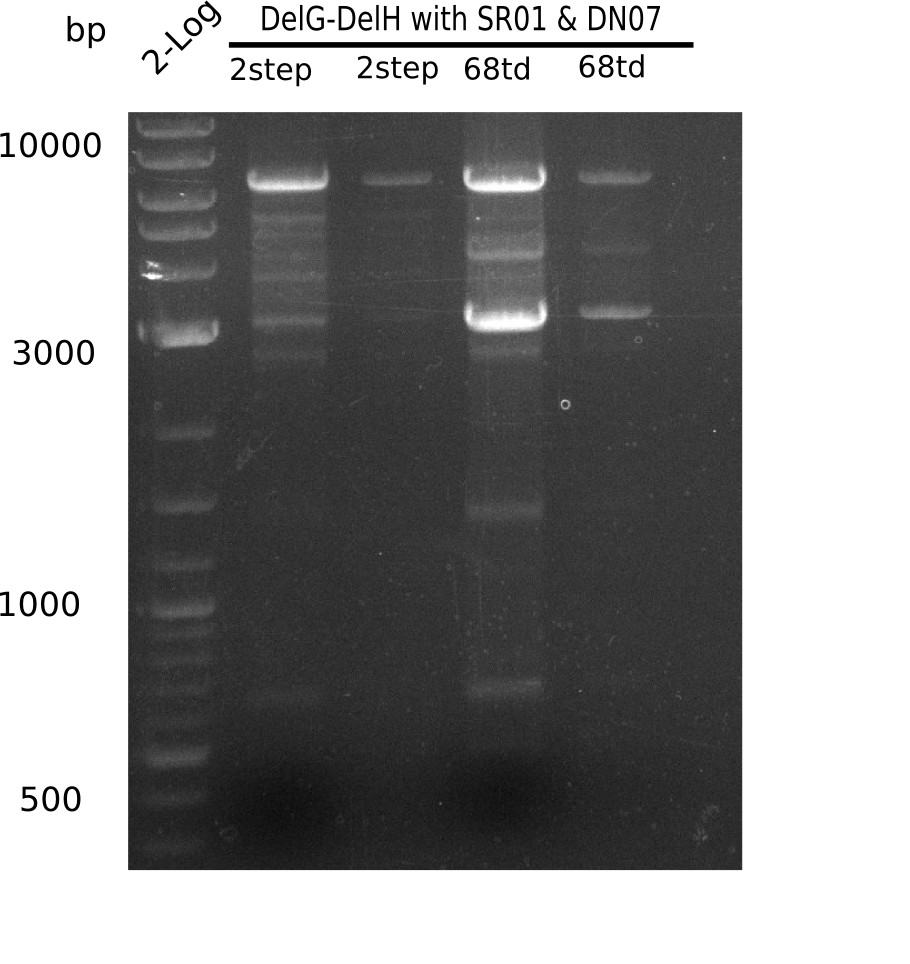
In week 7, we reamplified the DelH fragments used for the transformation in week 6. We found out that amplification of fragments F1a and F1b was not reproducible. As conclusion, we designed new primers for DelH, which are capable to amplify the beginning of DelH in a more efficient and specific manner. The overview summarizes the primers we ordered and their performance in amplificating DelH F1a. Primer DN11 worked really well and is used in the next experiments.
Week 8
After improving the amplification of the beginning of DelH (fragment 1a with the primer DN11), we used a higher concentration in the ligation assembling the pHM01 plasmid. The transformation of DH10ß cells was performed with electroporation. Four colonies were positive in the screening-PCR, but did not express lacZ (no blue color). To quantify if DelH is possibly expressed anyhow, we plan a SDS-PAGE and induce the bacteria with higher concentrations of arabinose and X-Gal.
Week 9
The SDS-PAGE showed no clear band at ~600 kDa, so we conclude that there is most probably no DelH expression in the analyzed colonies. To confirm the result, a restriction digest was performed. The colonies did not show the expected pattern, so they indeed did not contain the desired plasmid. To perform a new assembly of the plasmid pHM01, the fragments of DelH and the backbone had to be reamplified. The amplifications of the DelH fragments went well, but the backbone caused again some difficulties and it was not possible to amplify it.
Week 10
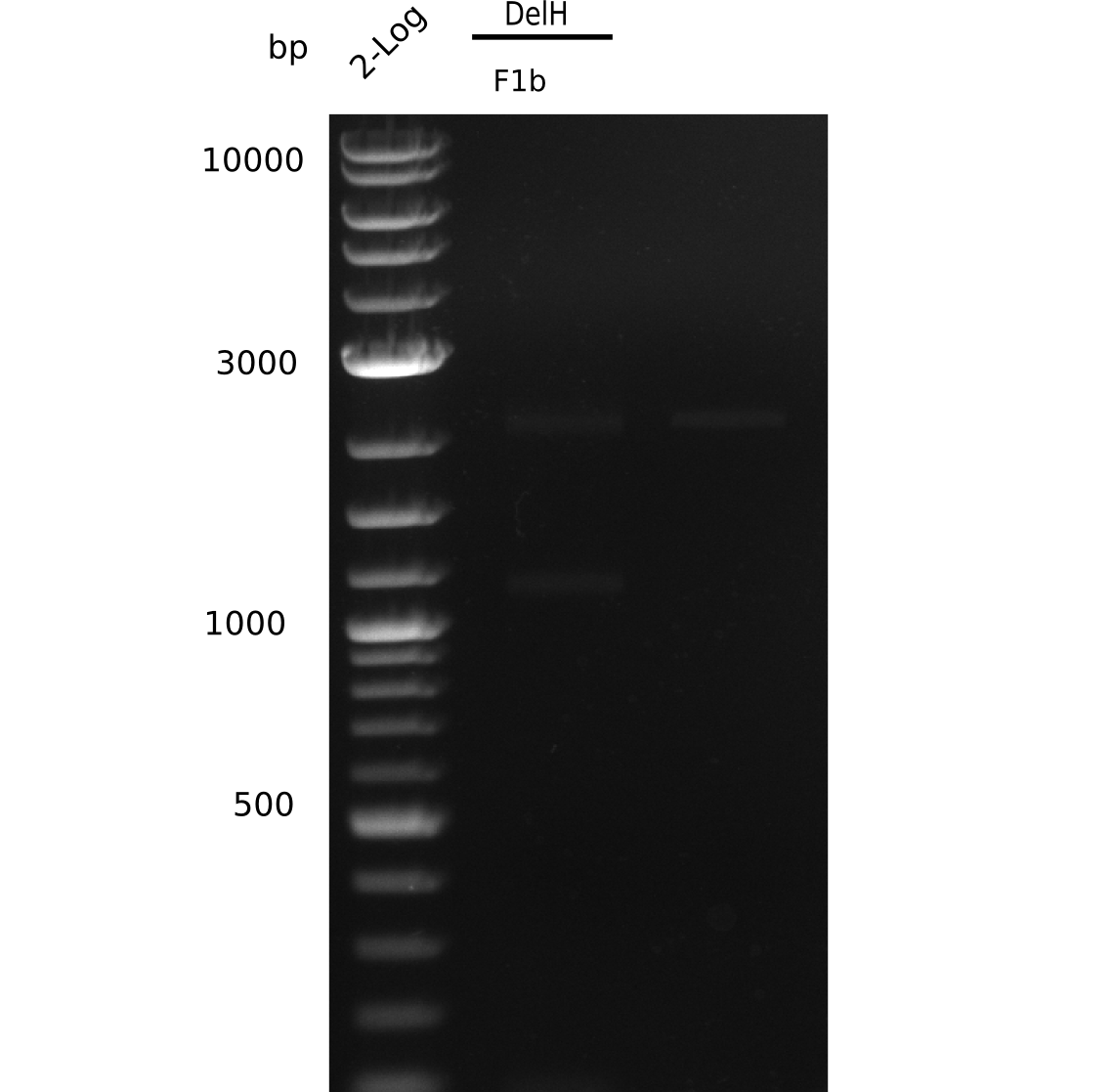
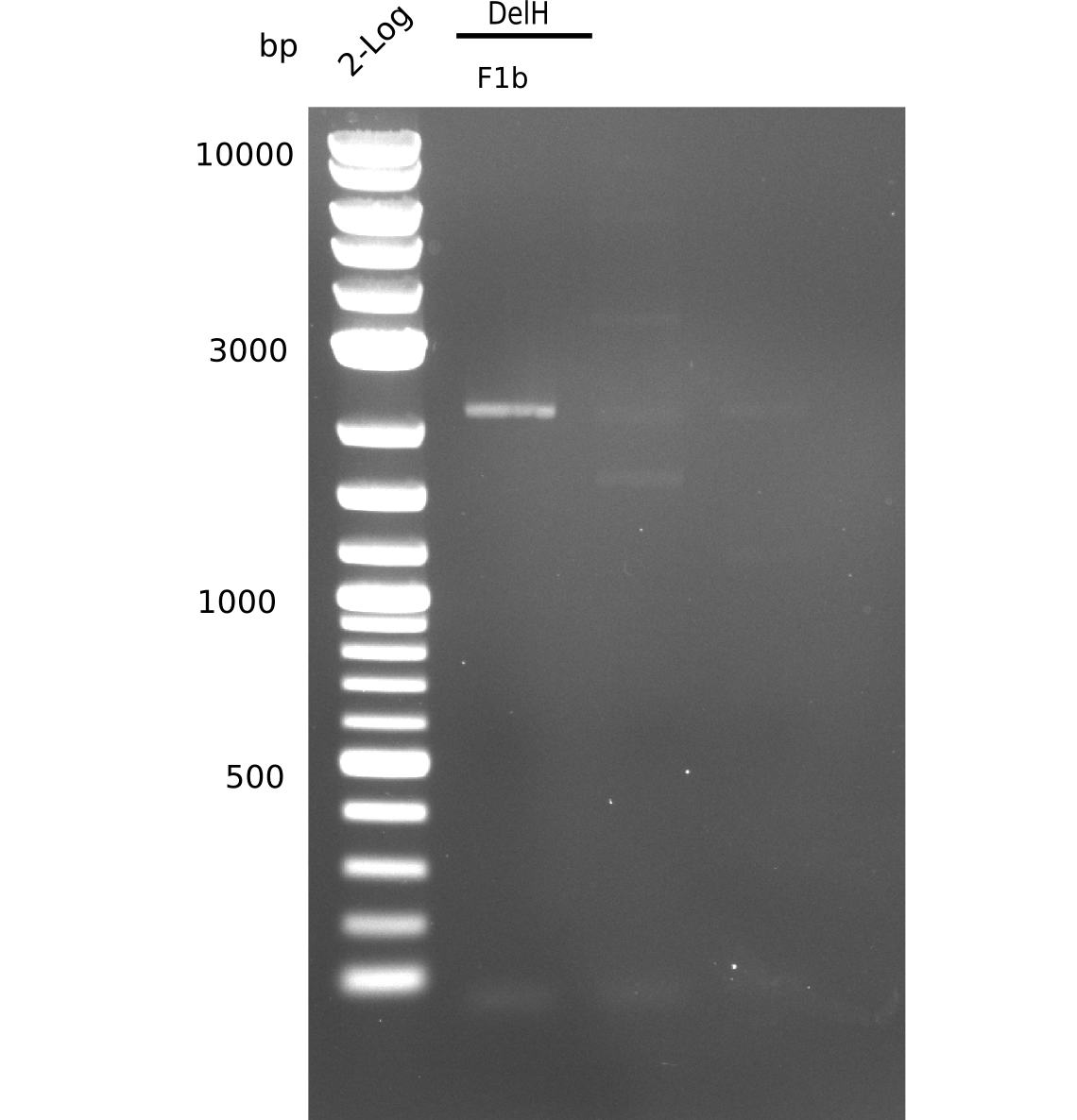
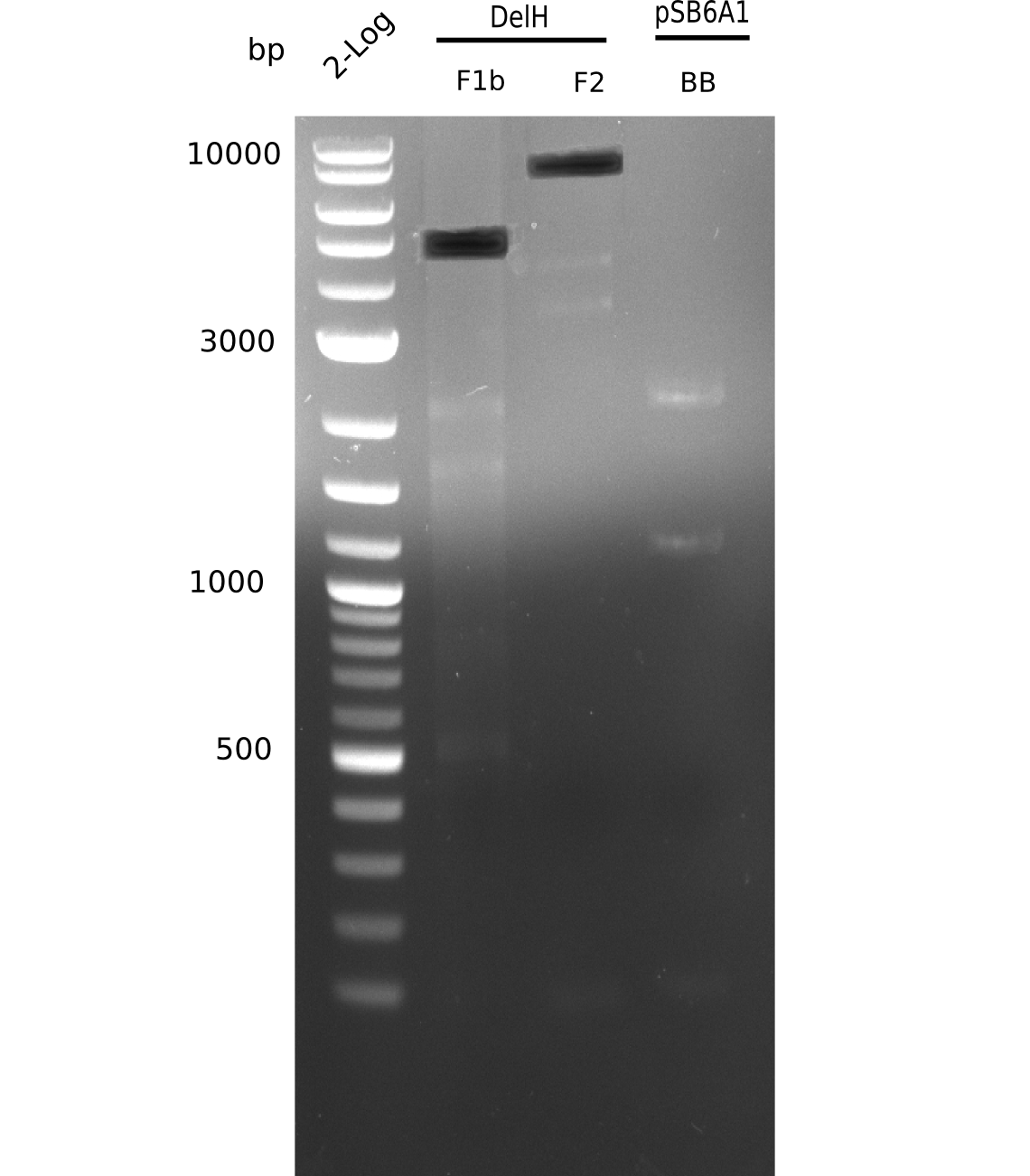
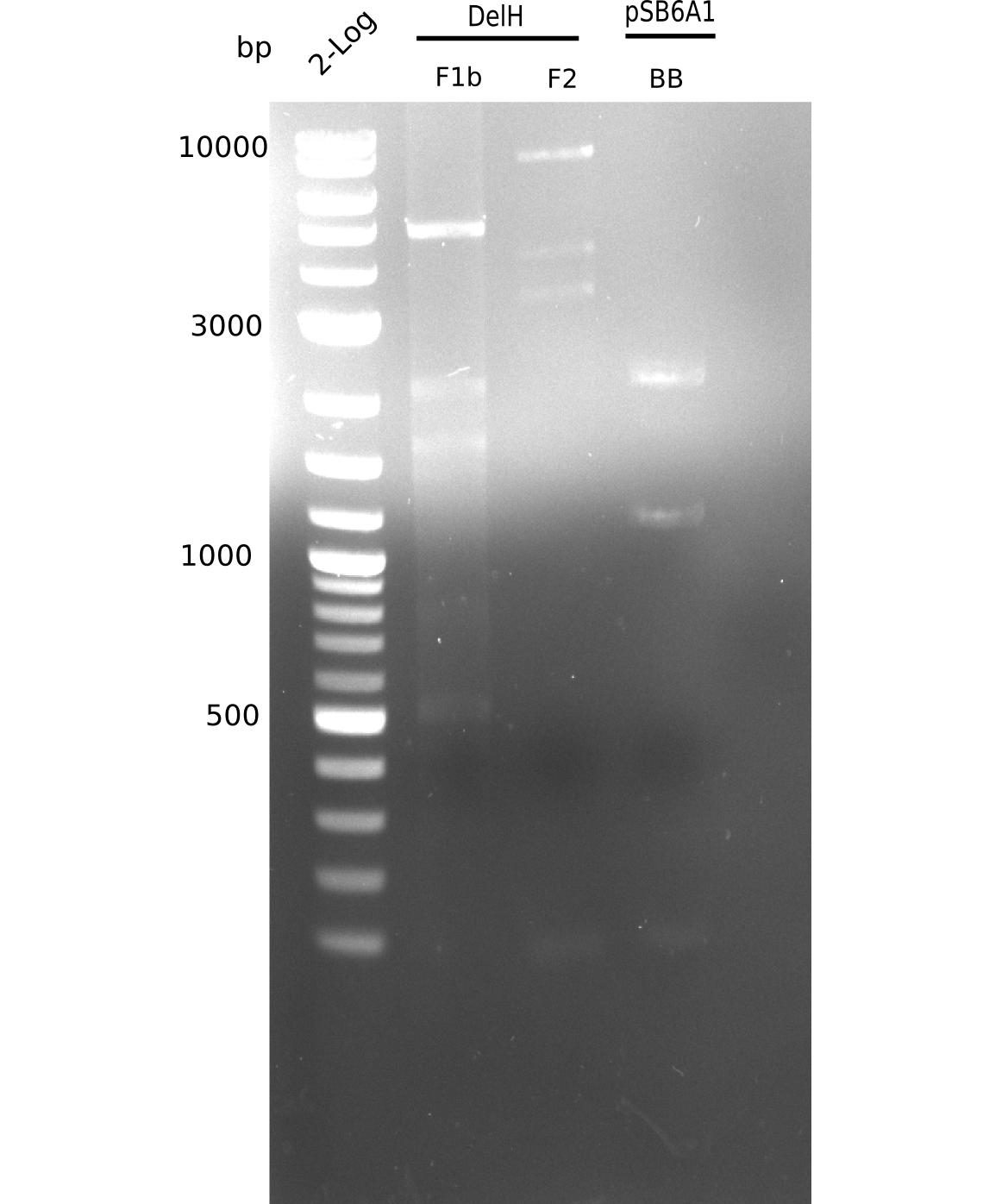
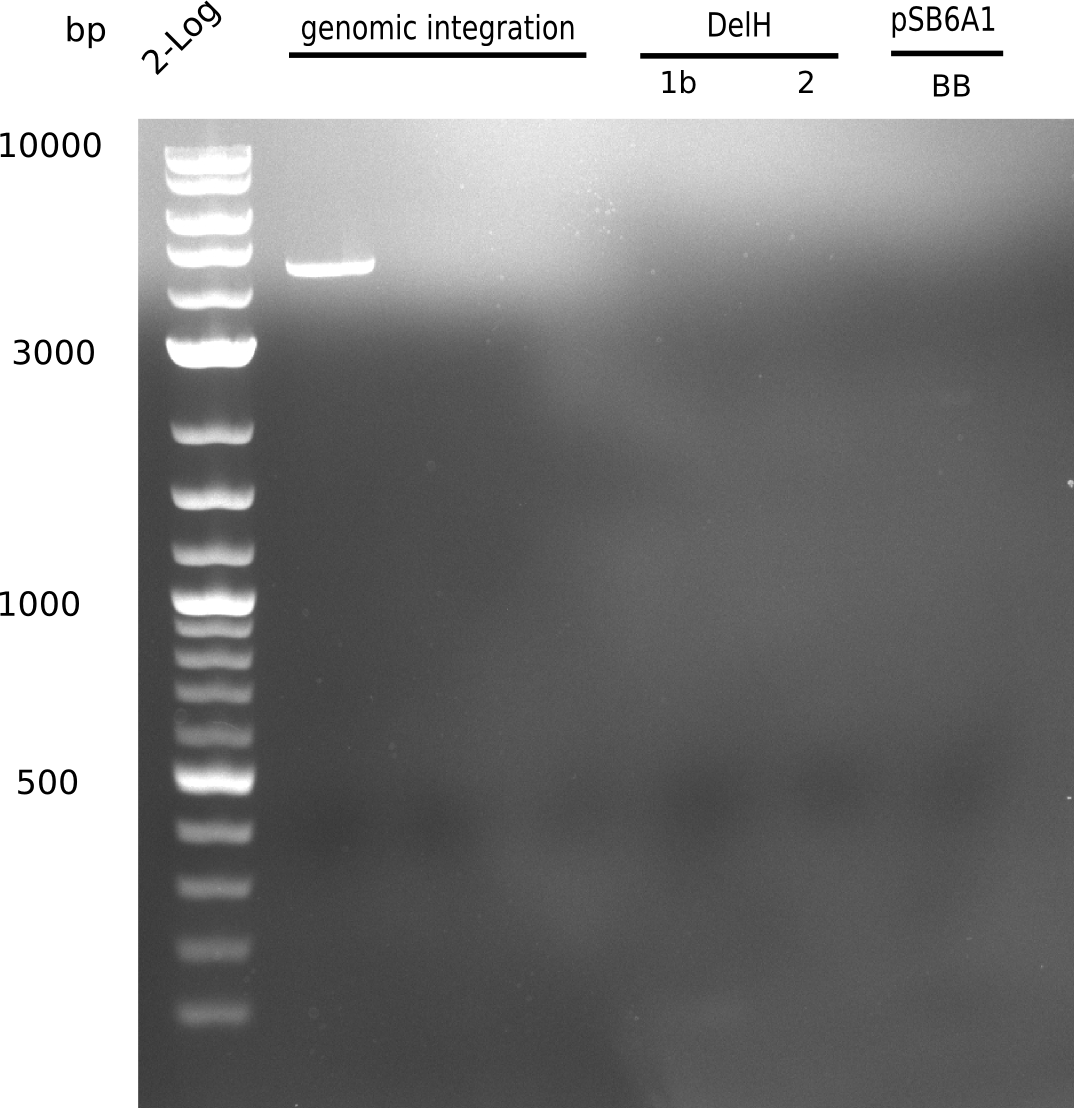
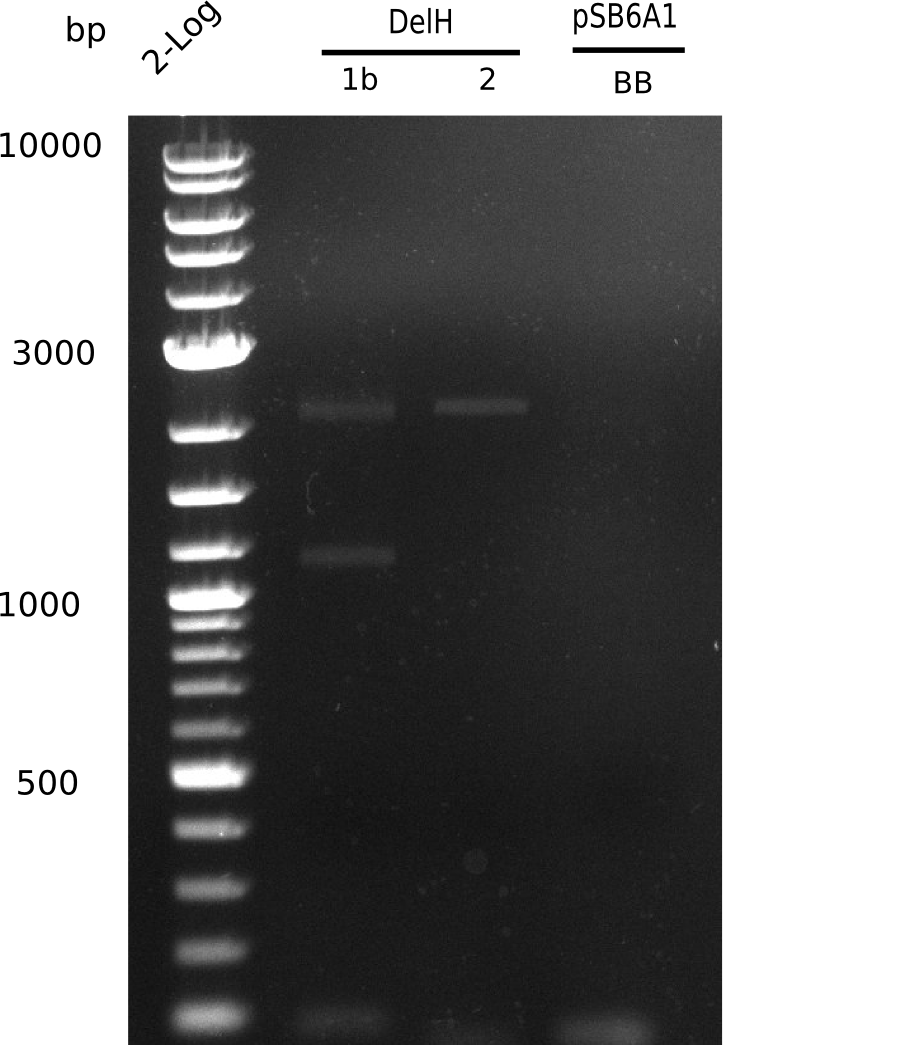
When rechecking all amplified fragments, we did not find what we expected. On the one hand, the concentrations of the fragments were too low and could not be seen on the gel. On the other hand, a re-amplification of the fragments with the appropriate primers was not successful. Additionally, a new polymerase (Phusion Flash) was used. Interestingly for the DelH 1b fragment, the fragment amplified from the genomic DNA was slightly larger than the amplified fragments from itself.
Week 11
Based on this week's experiments and in silico analysis to validate the fragments DelH F1a, F1b and F2, we discarded the restriction digest and ligation strategy for the assembly of the DelH plasmid. Instead, we chose Gibson assembly as alternative method, developed a strategy and designed primers accordingly. We are positive, to finally assemble pHM02.
Week 12
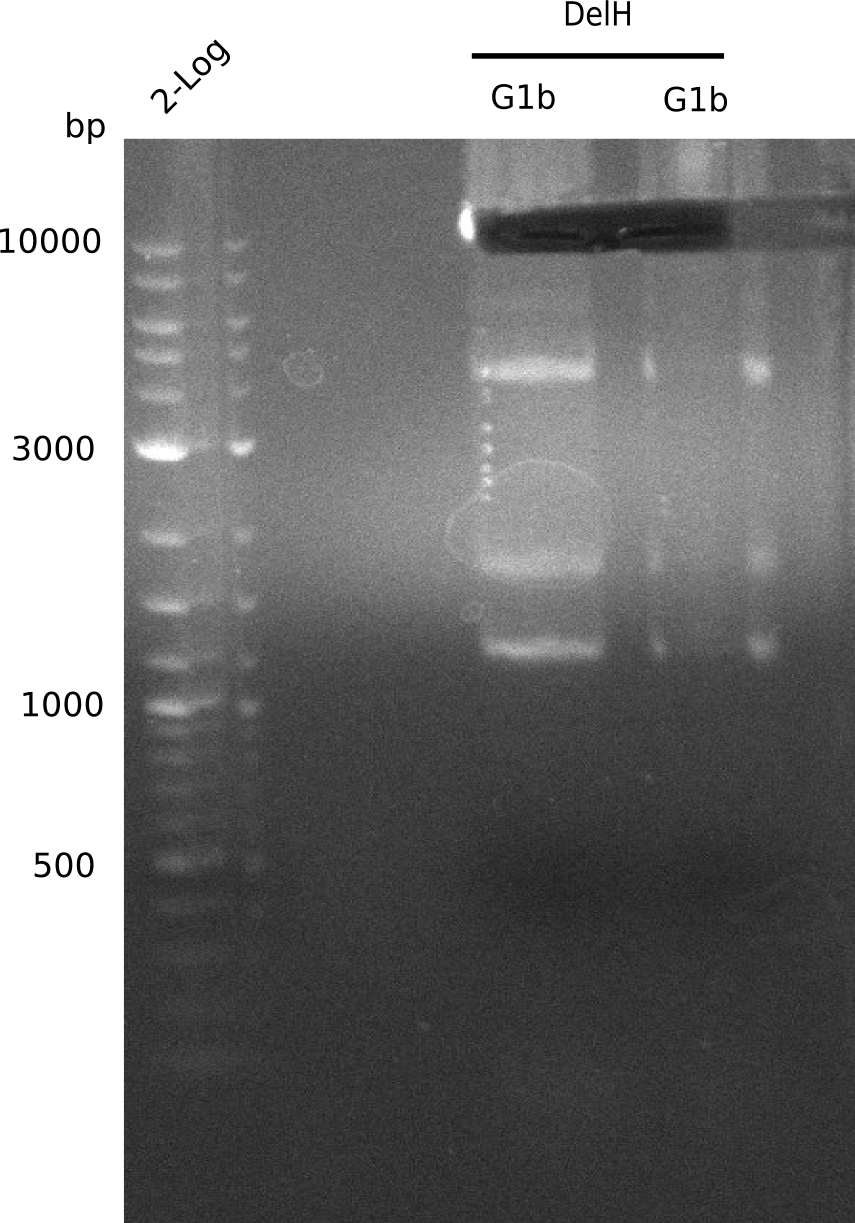
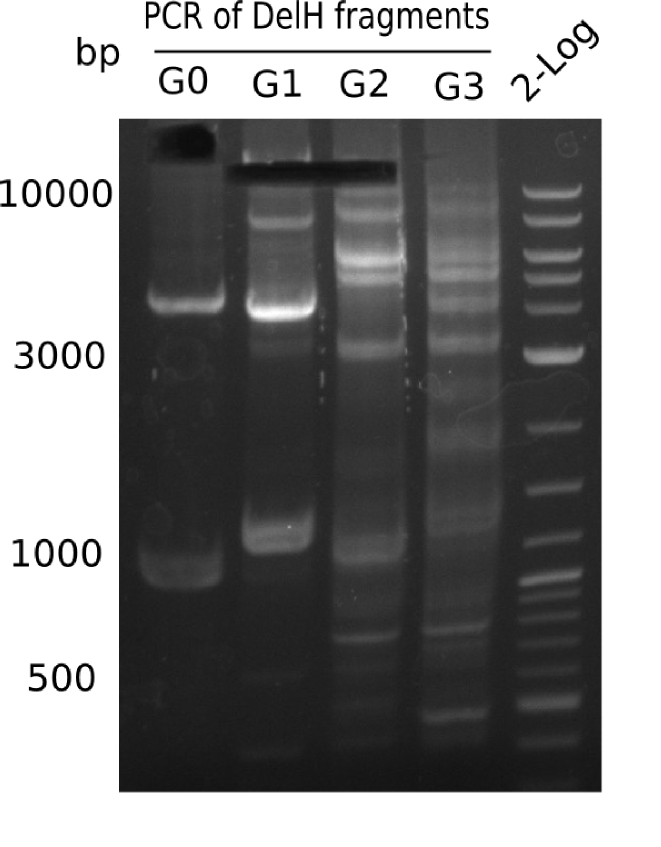
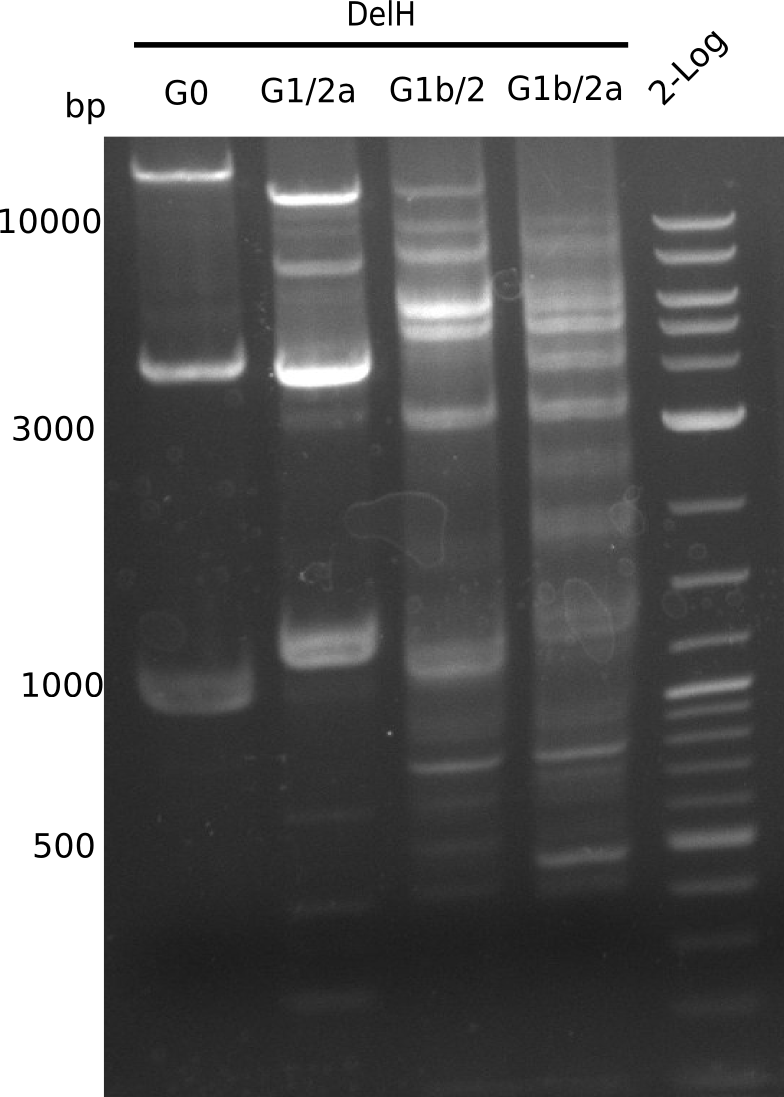
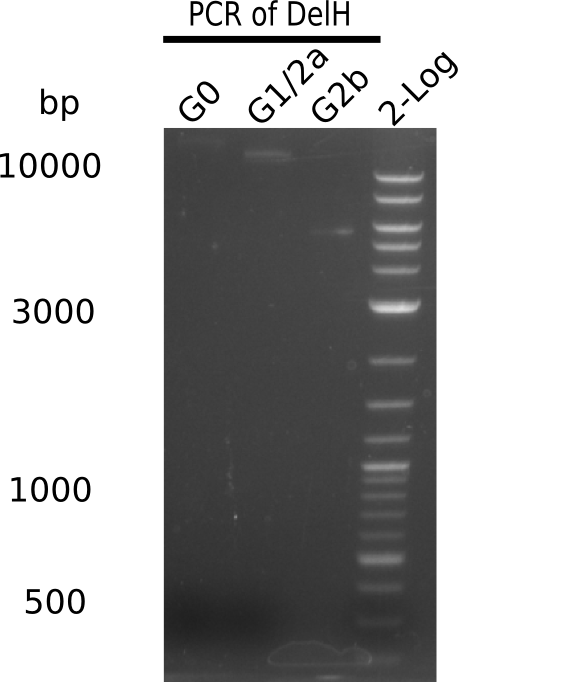
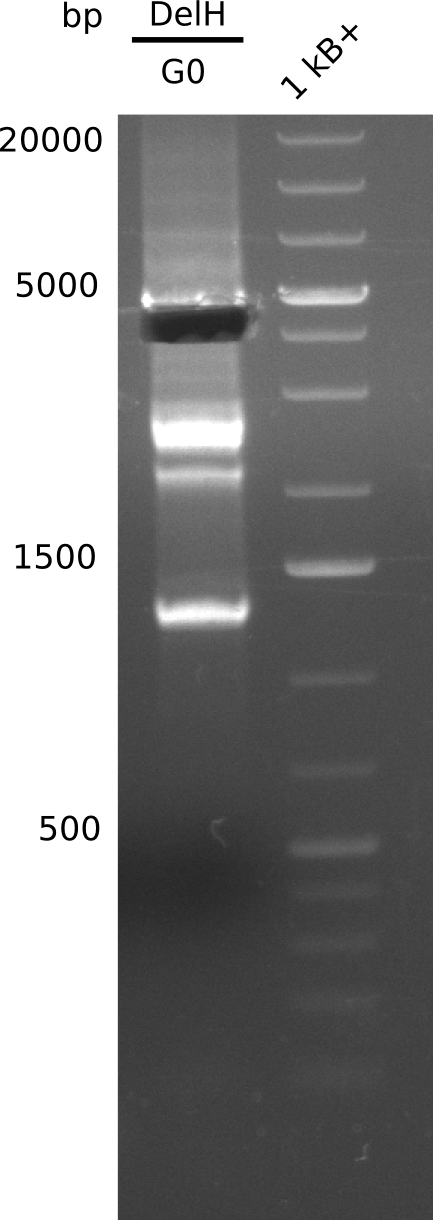
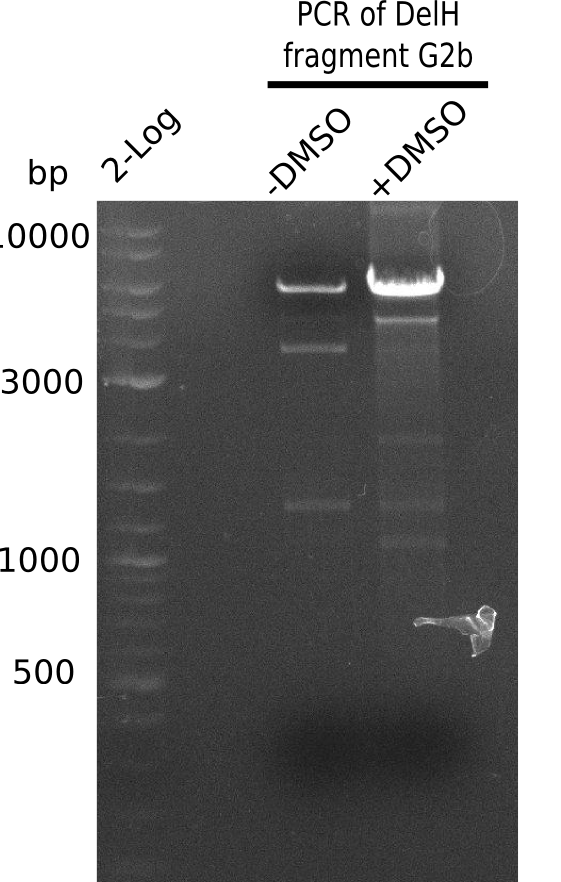
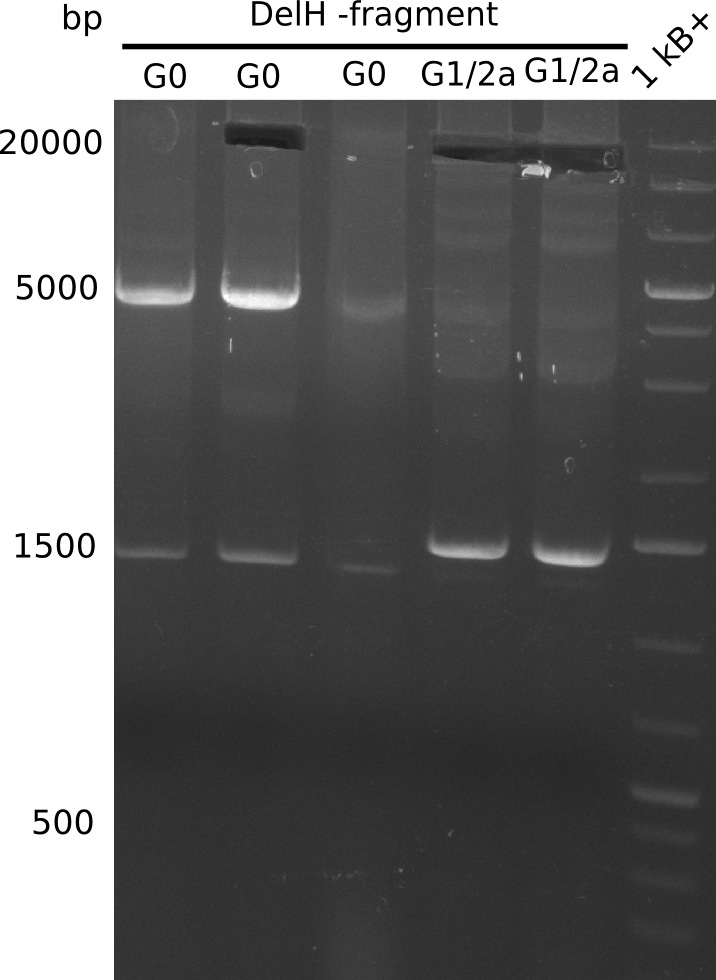
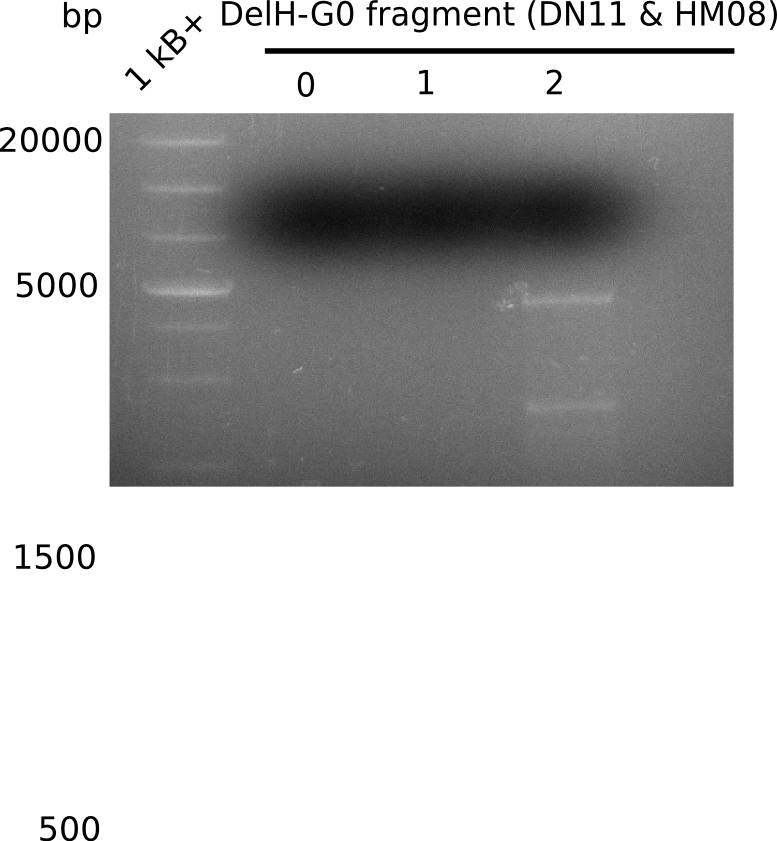
The primers arrived and we started amplifying the Gibson fragments using different approaches. Since G1 could not be PCR amplified, we decided to divide G1 into sub fragments G1a and G1b, for which we had designed and ordered primers already. The amplification of fragment G2 worked well and was produced in reasonable amounts. Alternatively, we also produced the entire DelH as one fragment G0, as well as further sub fragments DelH G1/2a, G1b/2 and G2b. Amplification of G1b/2a did not work out. Due to the failure of the restriction digest strategy, we further analyzed the backbone pSB6A1-AraC-lacZ, which we decided to discard and instead, use pSB6A1 and BBa_J04450, which is already available in the parts registry.
Week 13
In order to realize the new strategy using the already existing backbone from the parts registry pSB6A1-lacZ-mRFP, we designed two new primers for the backbone amplification and created a map of pHM03. The plasmid for the backbone was obtained from the registry, transformed in E.coli and minipreped. The Gibson fragment of the backbone was successfully amplified, together with Gibson fragments DelH G0, G1 and G2b.
Week 14
In week 14, we've tested numerous colonies from last week's Gibson assembly 28-07 using DelH G0 as well as G1/2a and 2b by screening-PCR. None of the analyzed colonies carried a correct plasmid. We therefore performed another Gibson assembly 01-08 and again screened numerous clones. We found few possibly correct ones that will be further analyzed next week. Additionally, the new D. acidovorans strain SPH1, whose sequence is available in GenBank, was ordered. We will have to amplify all fragments from this strain again as soon as it arrives.
Week 15
We further tested colonies from last weeks Gibson assemblies using DelH G0 as well as G1/2a and 2b. Unfortunately, they were all negative in the screening-PCR. Therefore, we amplified our Gibson fragments again and performed more Gibson assemblies. Yet again, there were no positive colonies in the colony-PCR.
Week 16
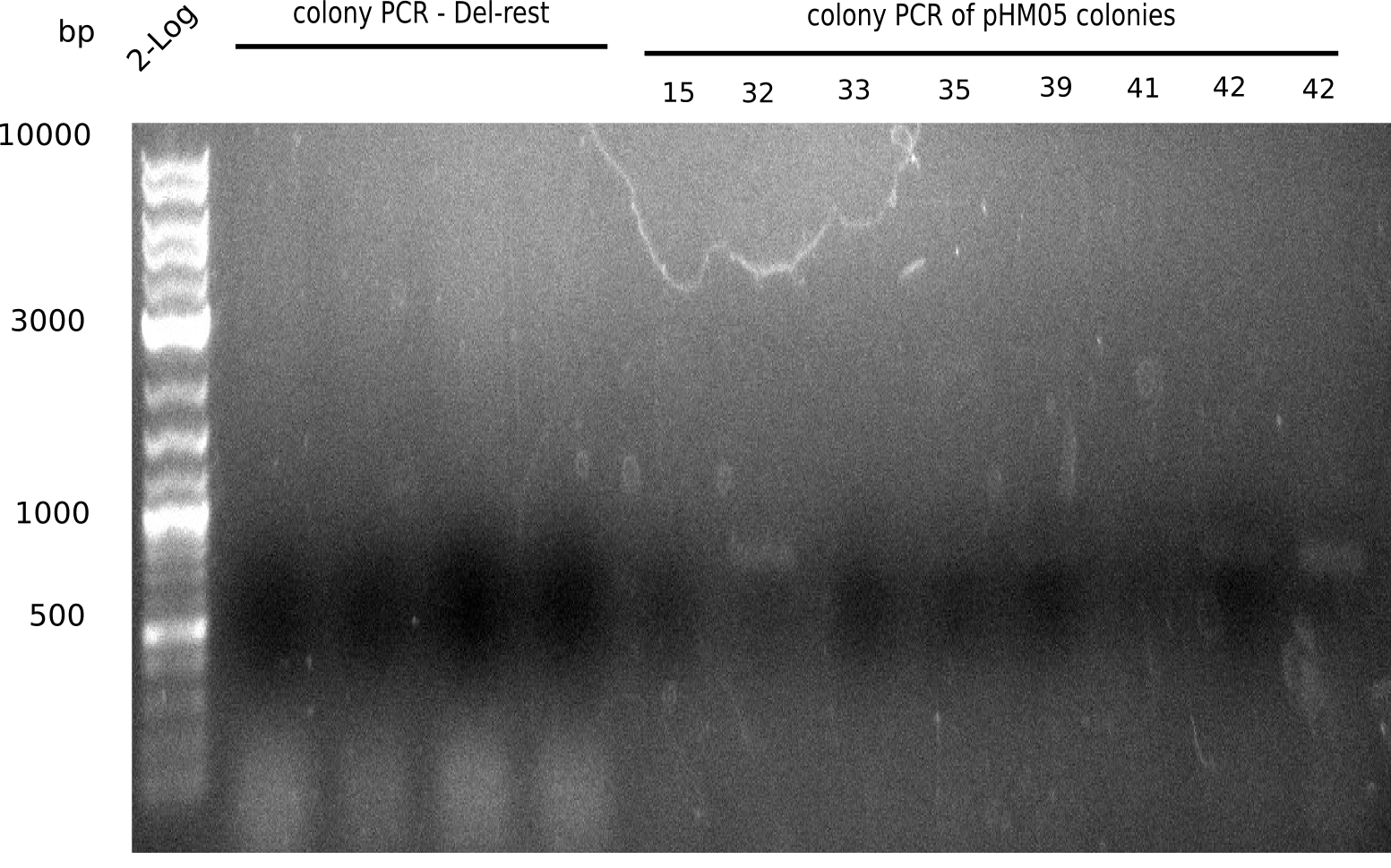
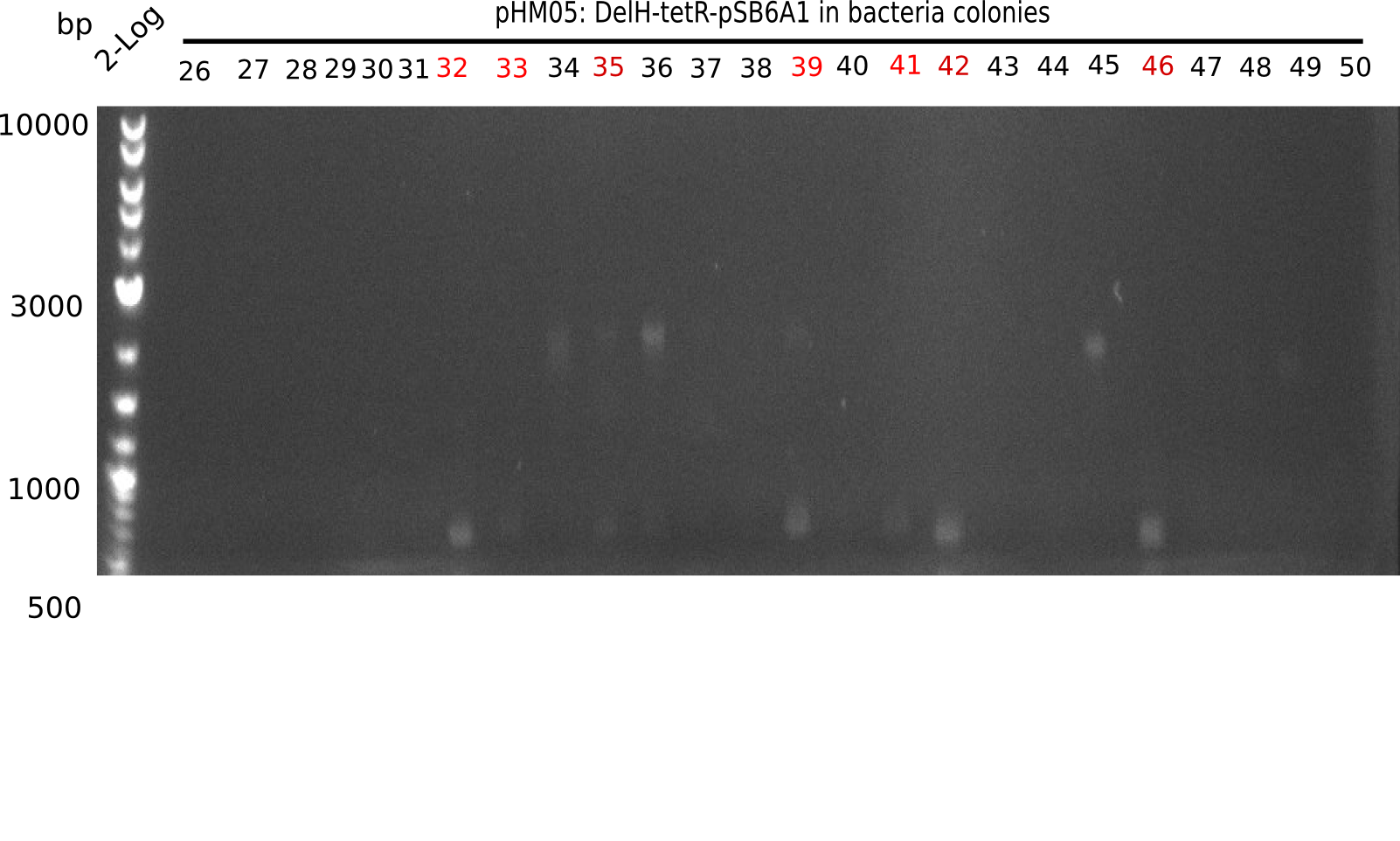
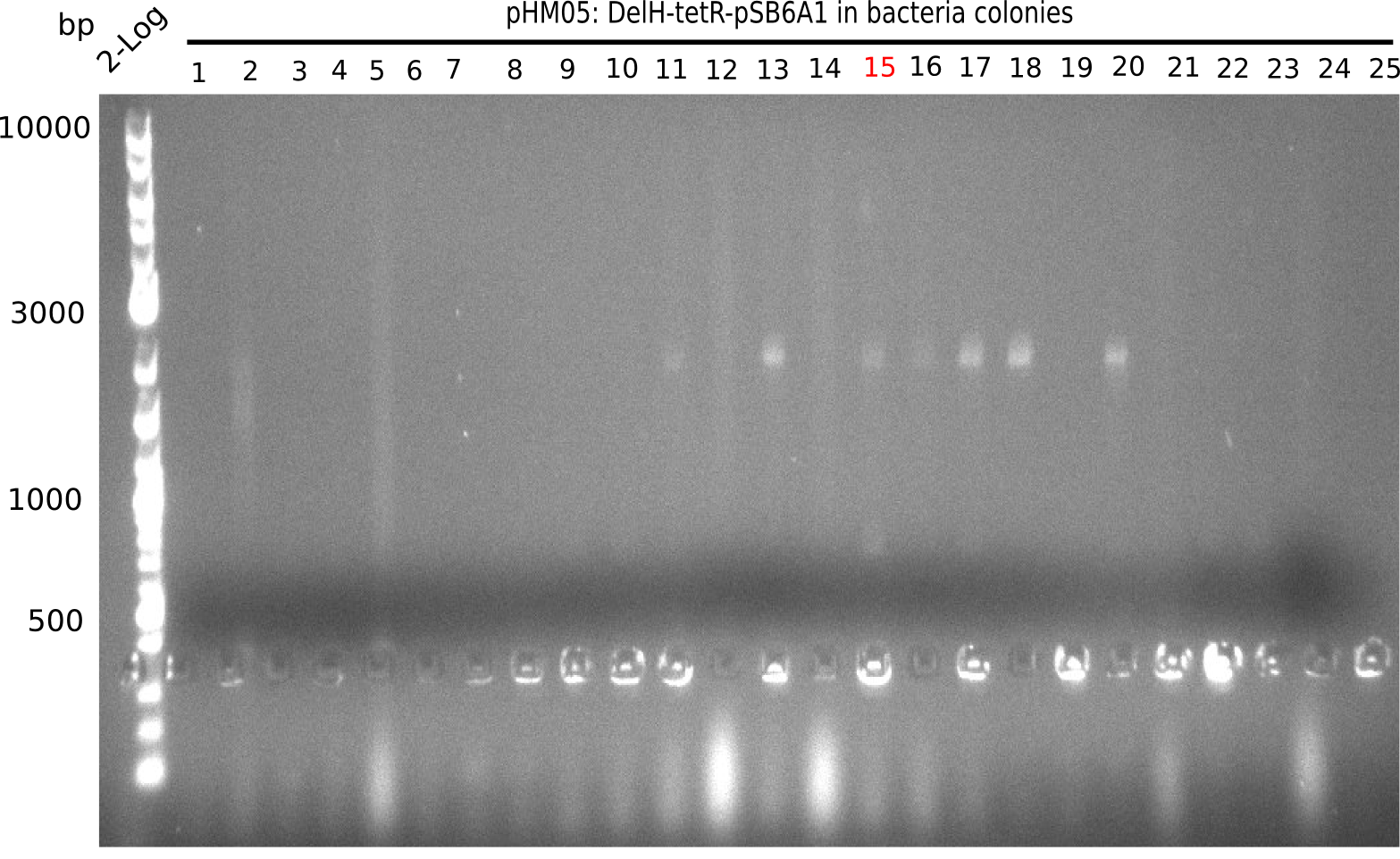
We further characterized the DelH plasmid created using Gibson assembly. Unfortunately, none of the screened colonies was for sure positive. Selection of red colonies was not clear, and PCR screened colonies were all negative. In order to avoid high background during screening of the colonies, we decided to run two different strategies. For first approach pHM04, we will amplify the backbone without mRFP, avoiding backbone reassembly due to ribosome binding site homology. In the second strategy pHM05, we will additionally introduce a tetracycline resistance, to ensure integration of the insert. In addition to the primers for the new strategies, we designed a new screening primer at the end of DelH.
Week 17
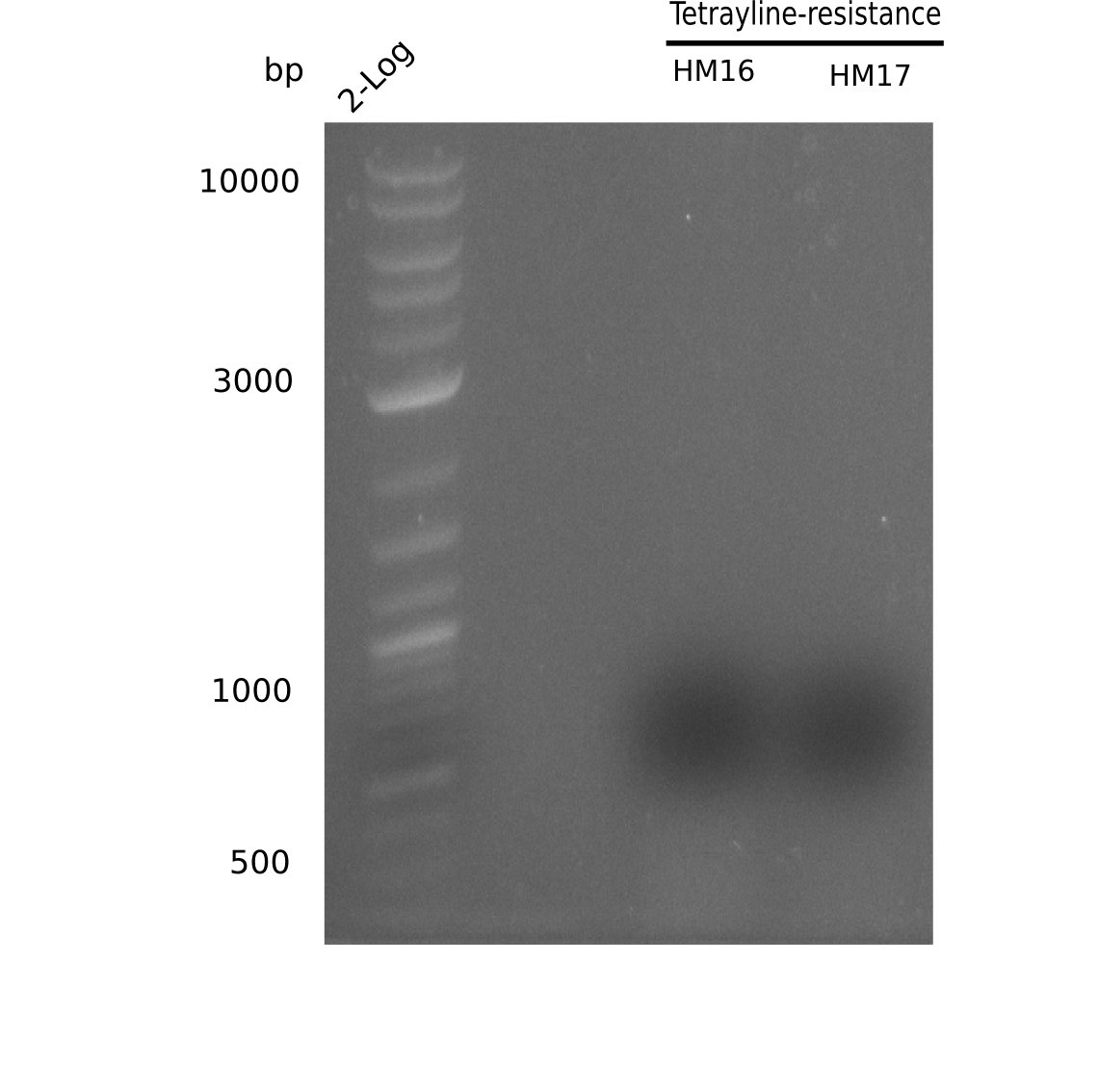
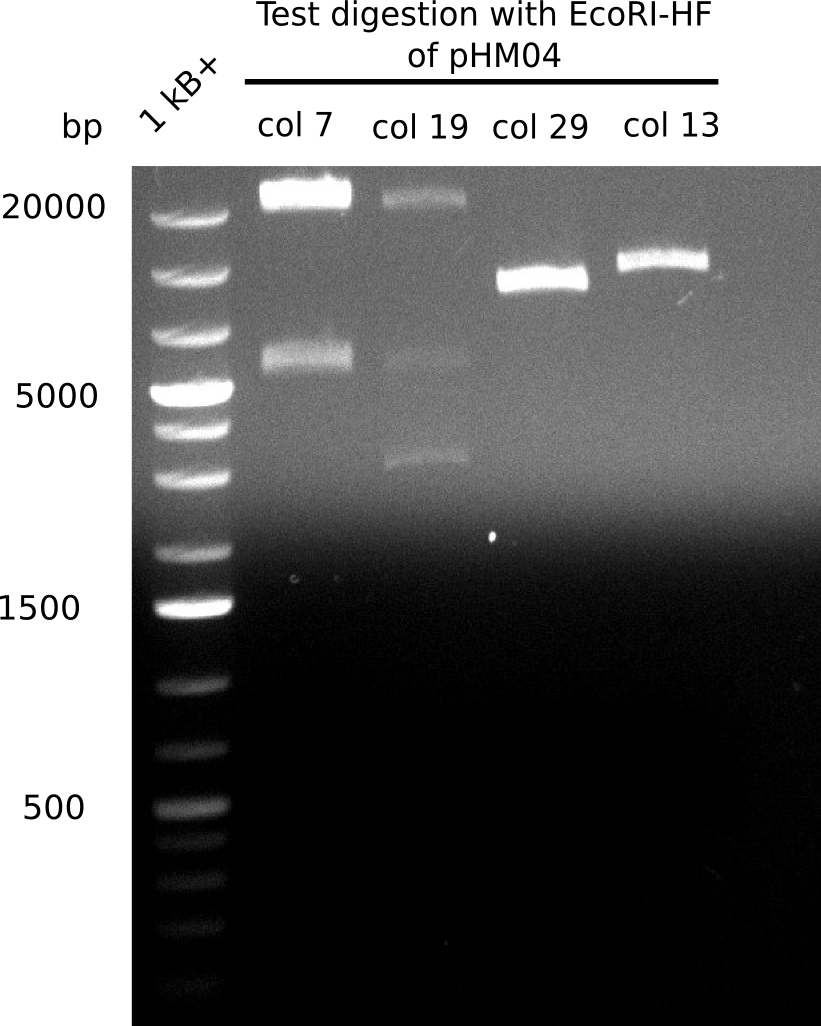
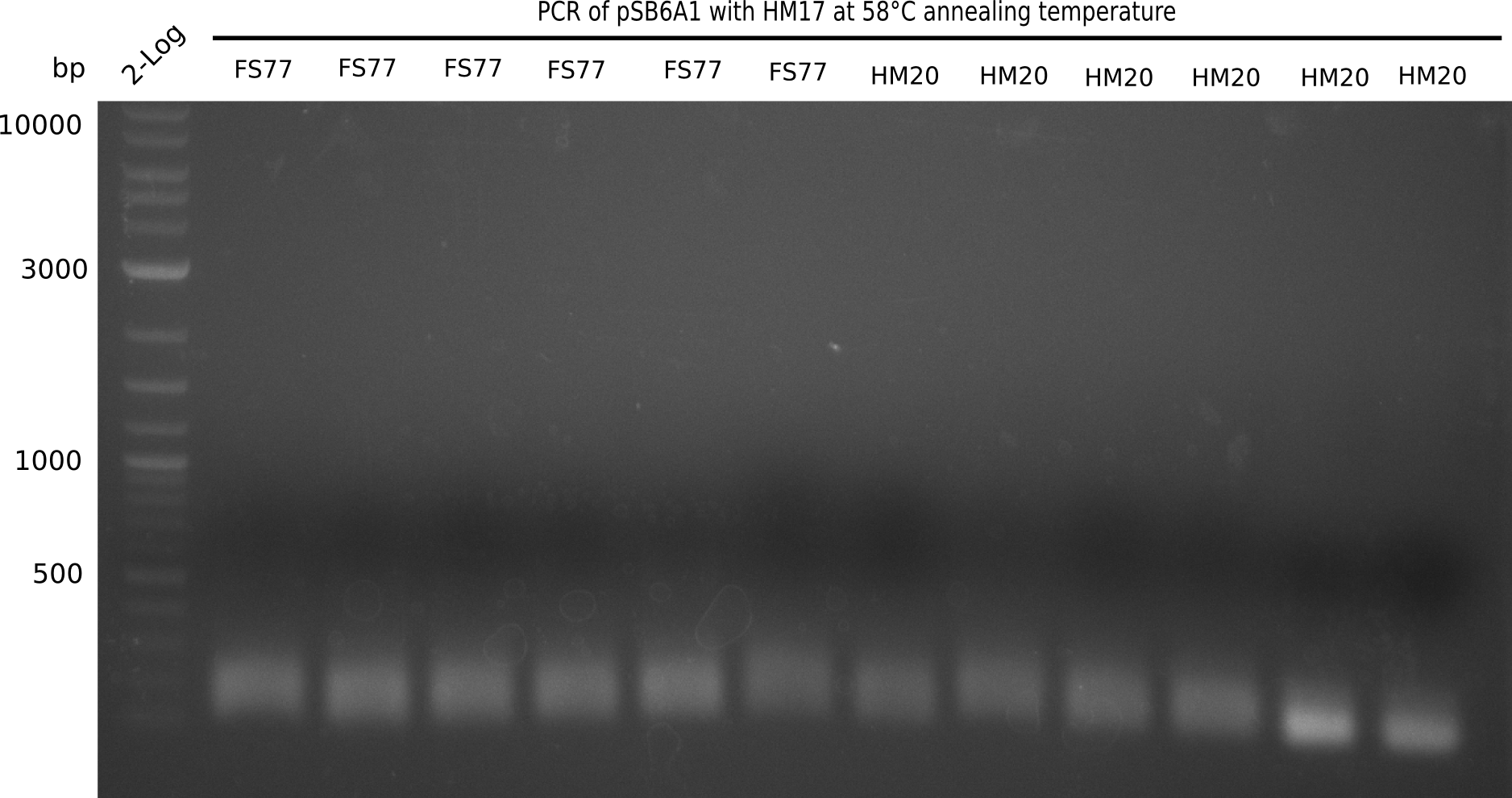
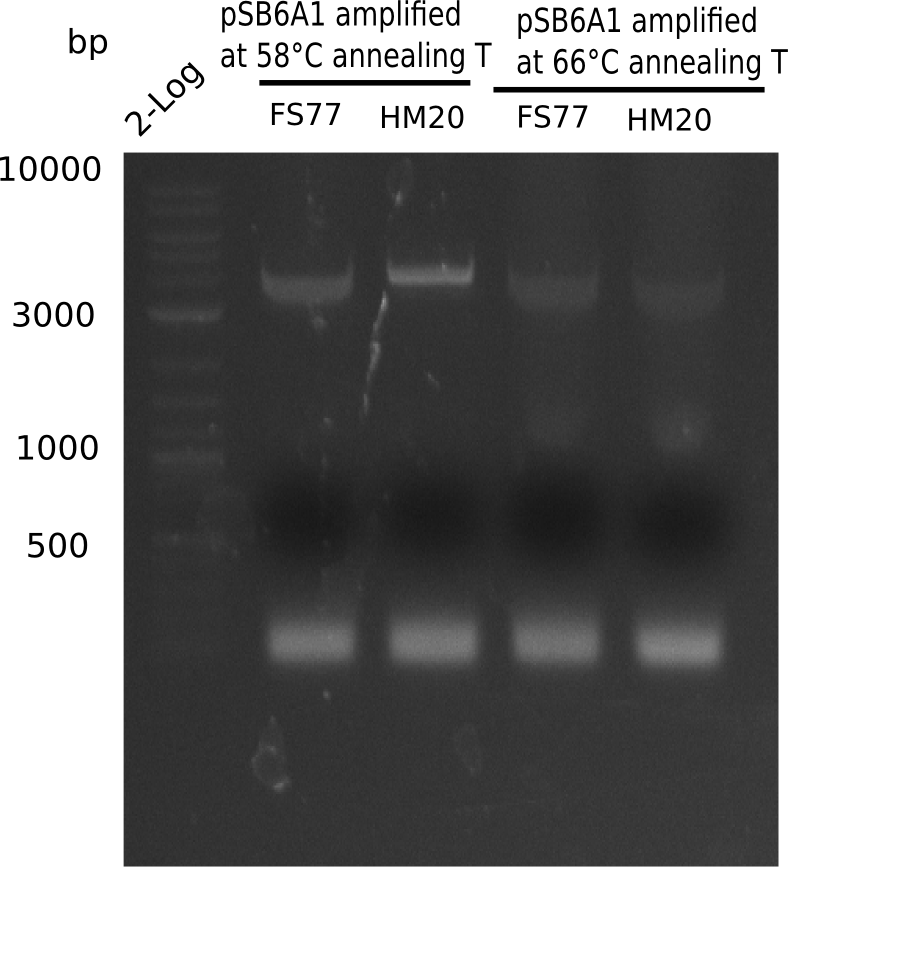
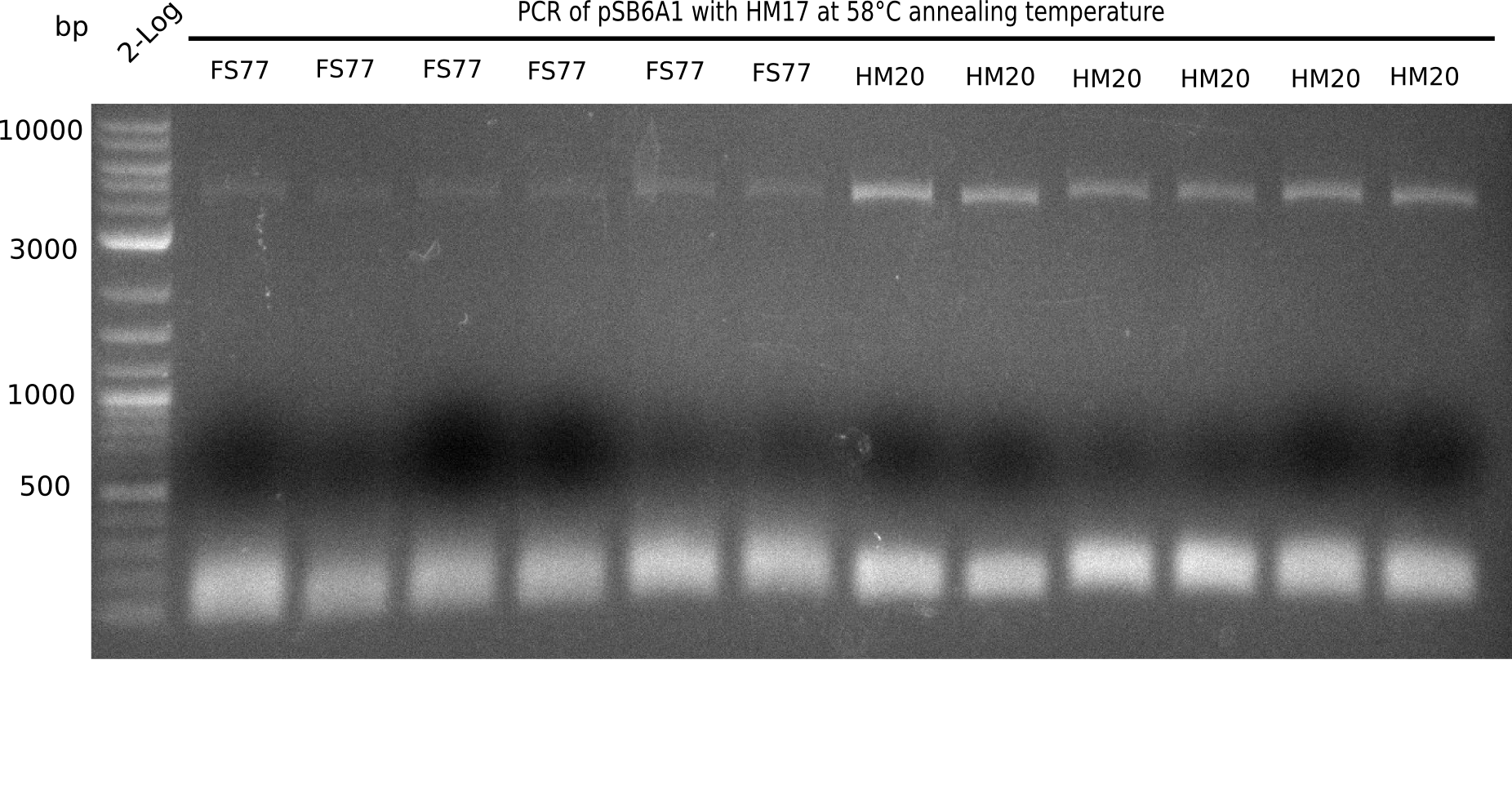
After performing different Gibson assemblies and screening always resulted in negative clones, the possible explanation could be a religation of the backbone fragment pSB6A1 allowing E.coli to survive and express mRFP. This week, our aim is to design a new construct without mRFP, so that we can exclude the red colonies from the screening. Therefore, we are going to use a new reverse primer for the backbone including only the terminator of the mRFP, but not the mRFP. The primers for the backbone are HM11 & HM17. The construct is named pHM04 and further explained in week 16.
Week 18
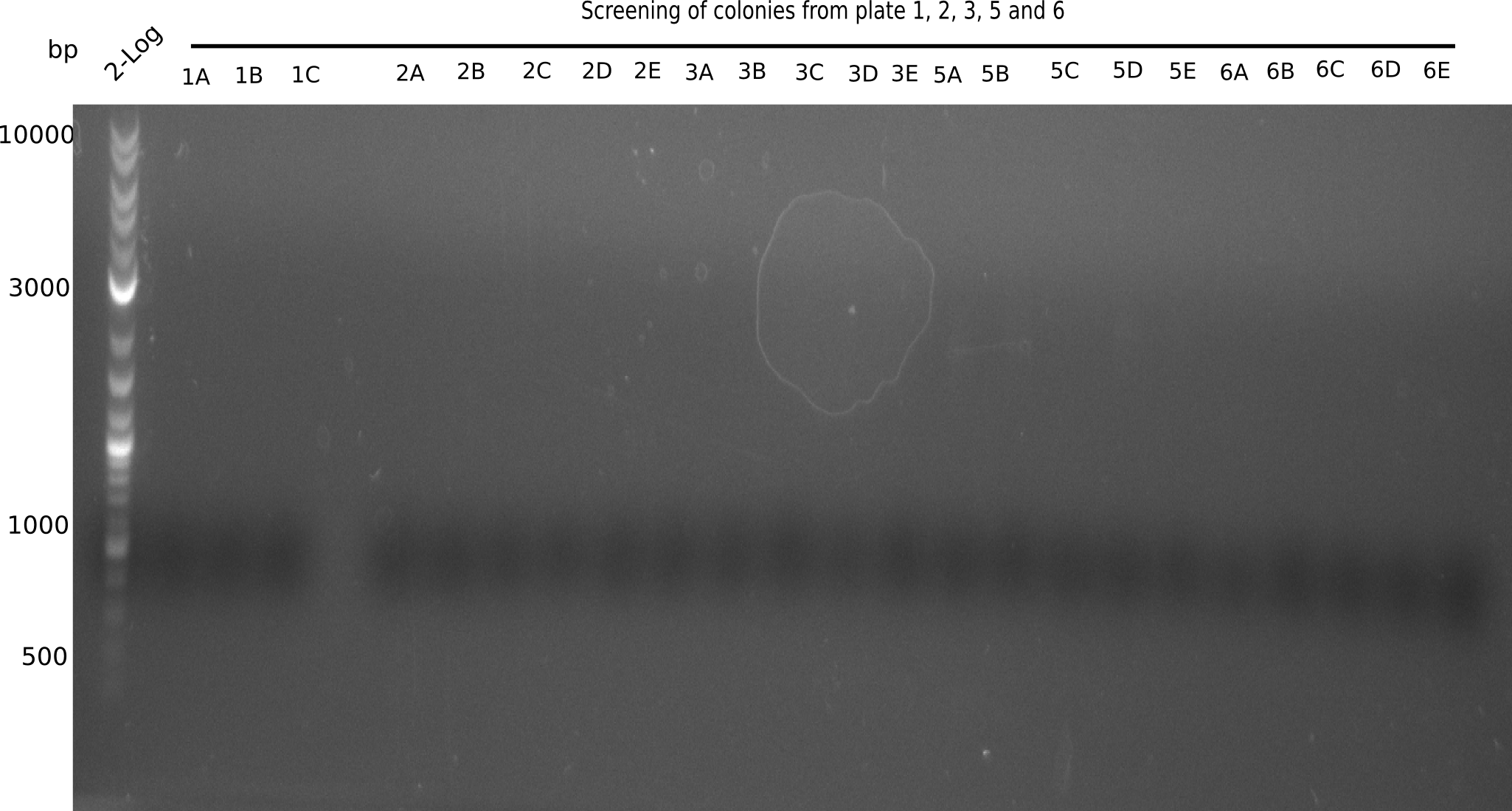
Because the constructing pHM04 did not result in any positive clone (see experiments week 17), we will follow the idea to introduce a tetracycline resistance to the ampicillin backbone as additional selection marker for successful assembly. So we have to screen only colonies, which are white (because of the exclusion of mRFP) and grow on plates containing tetracycline. Furthermore, we decided to amplify DelH in various fragments to increase Gibson assembly efficiency. Therefore, we also ordered new screening primers. The primers were designed as shown in the following table.
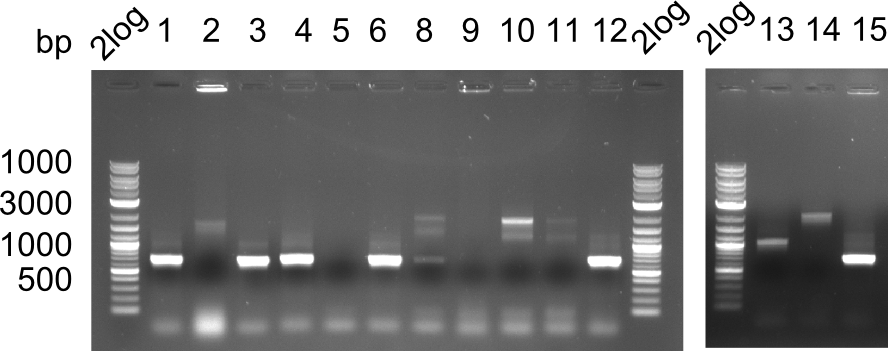
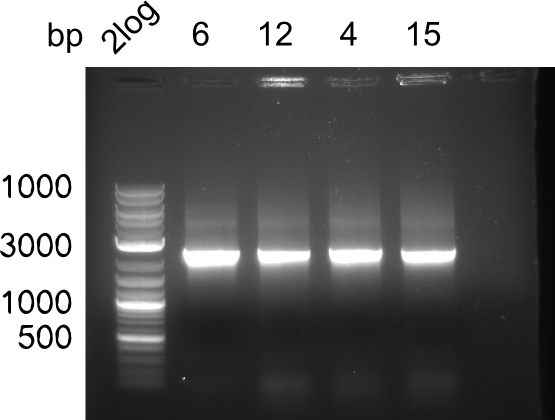
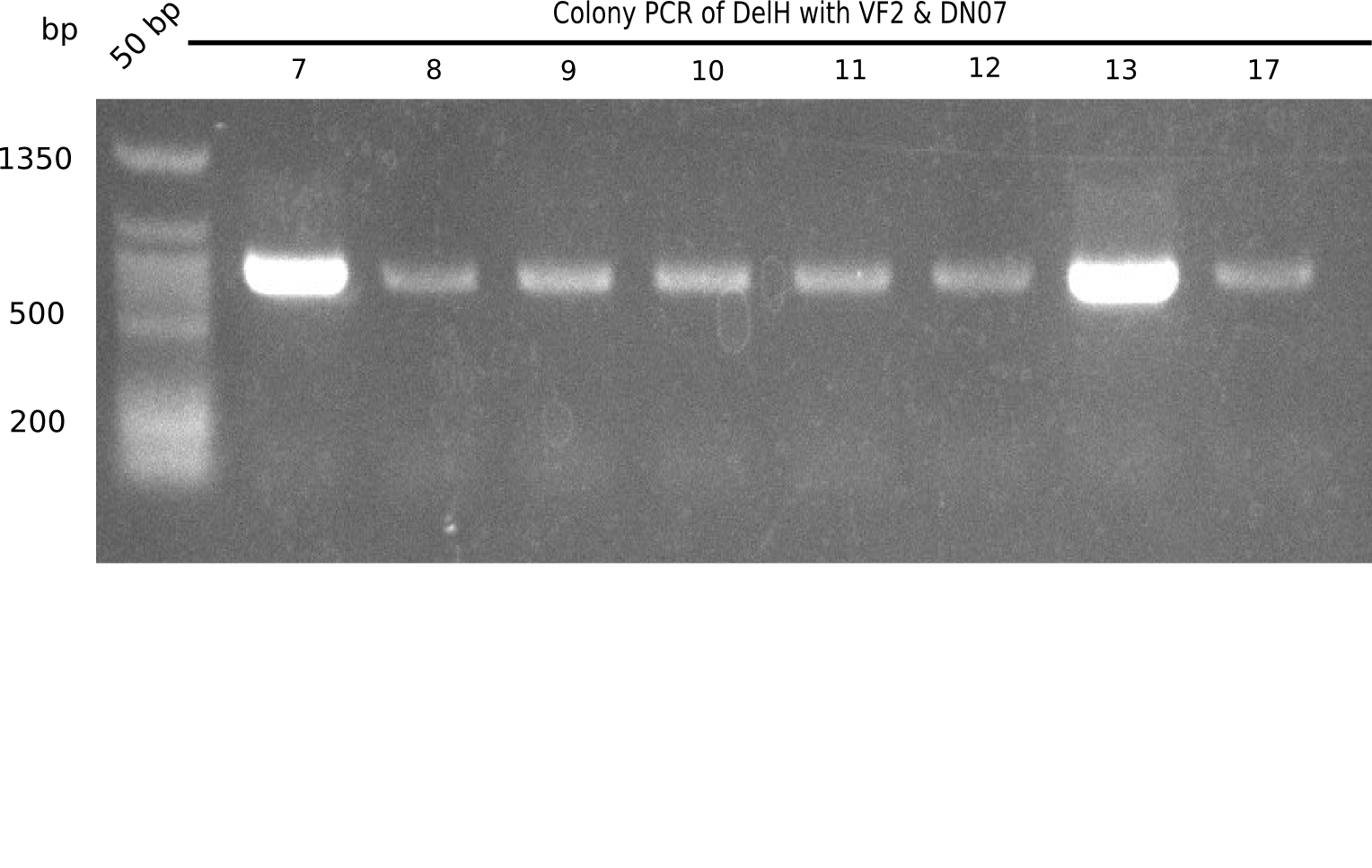
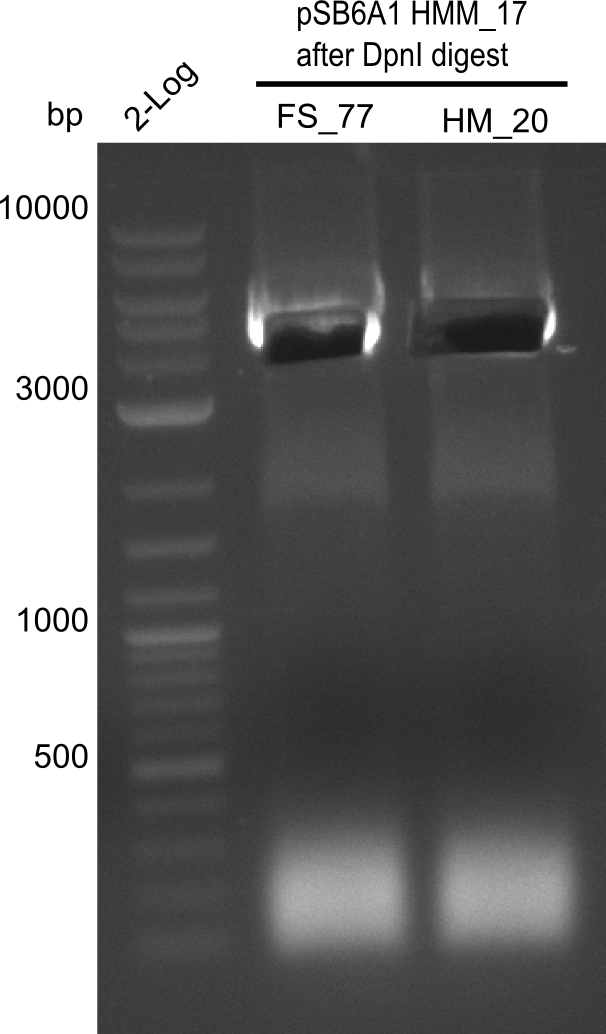
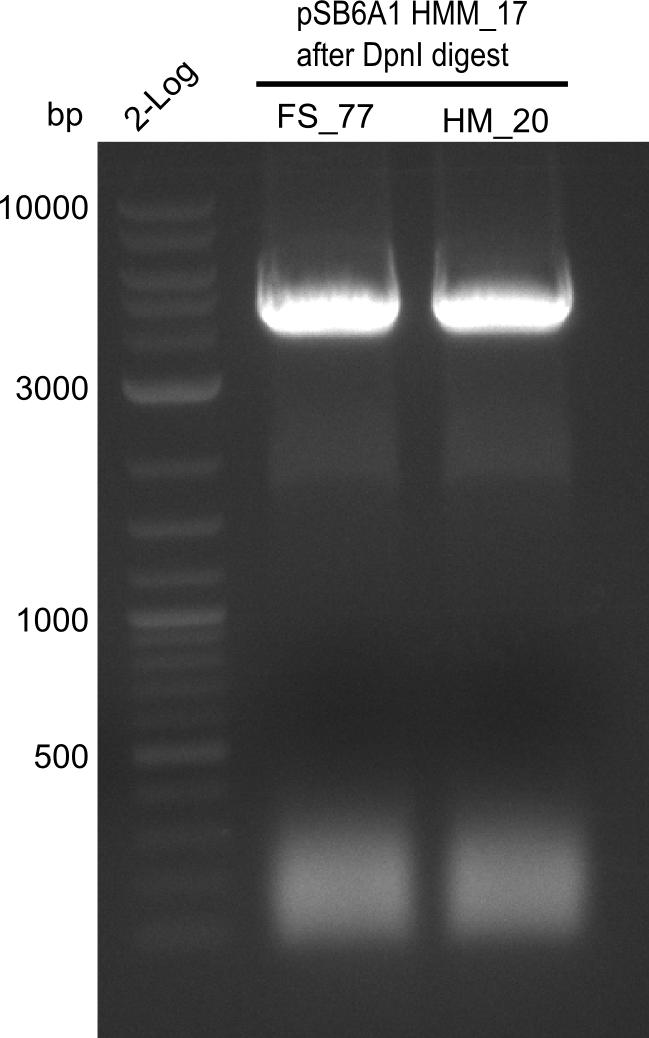
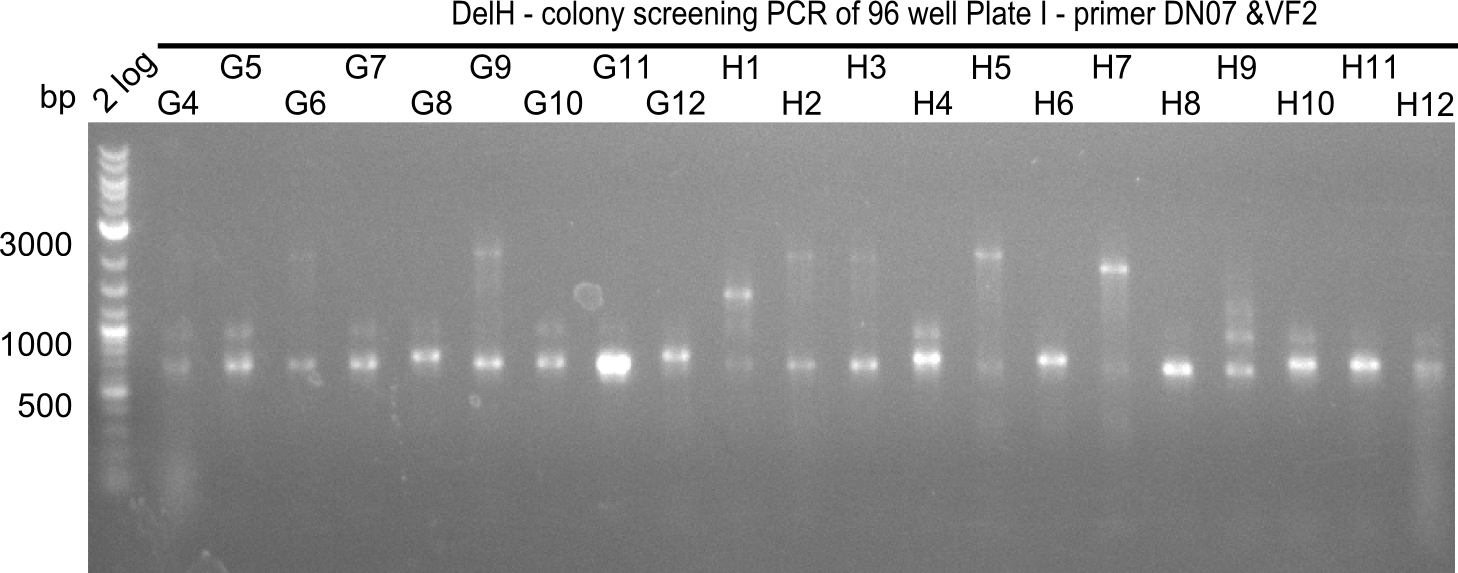
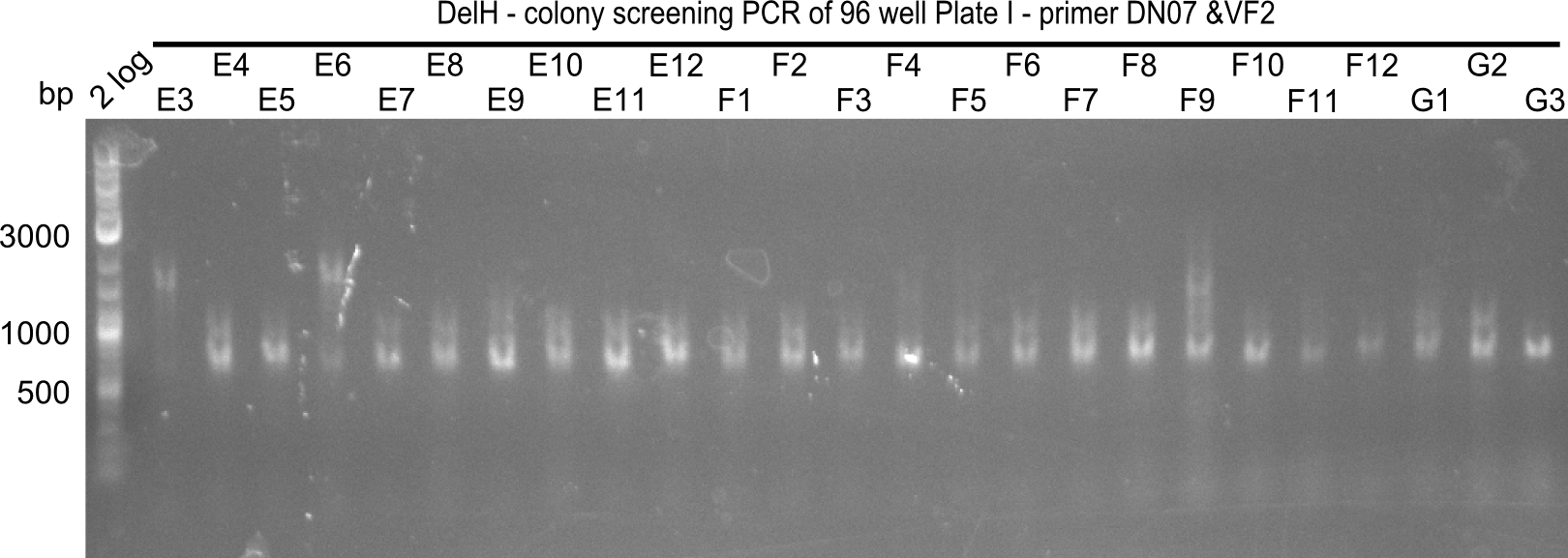
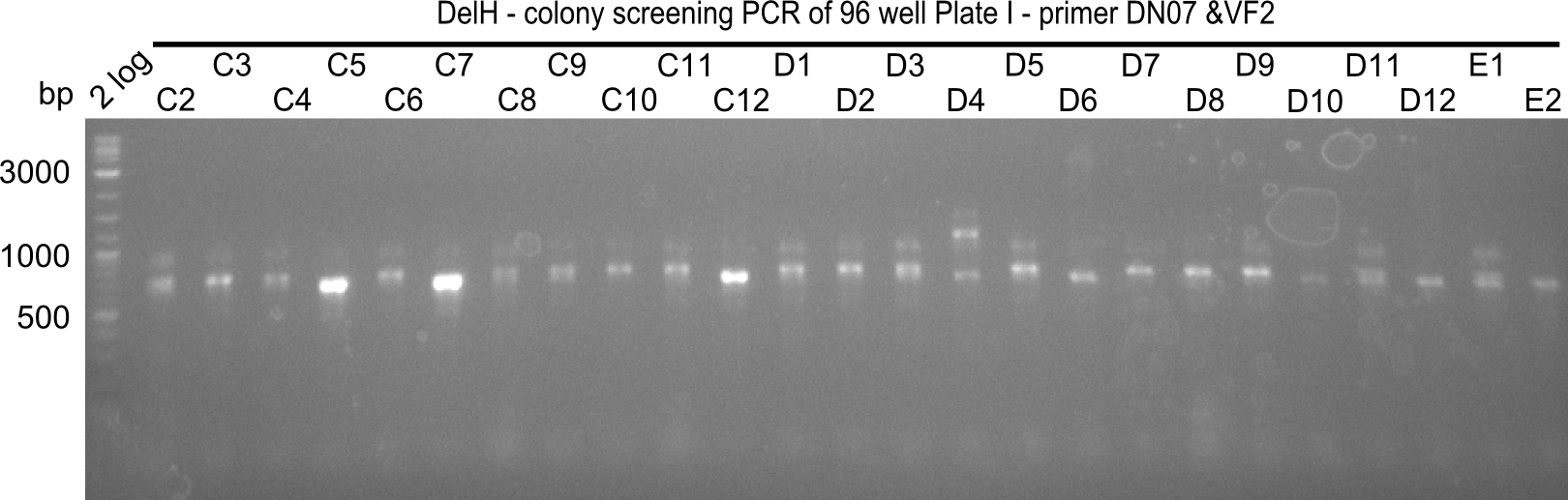
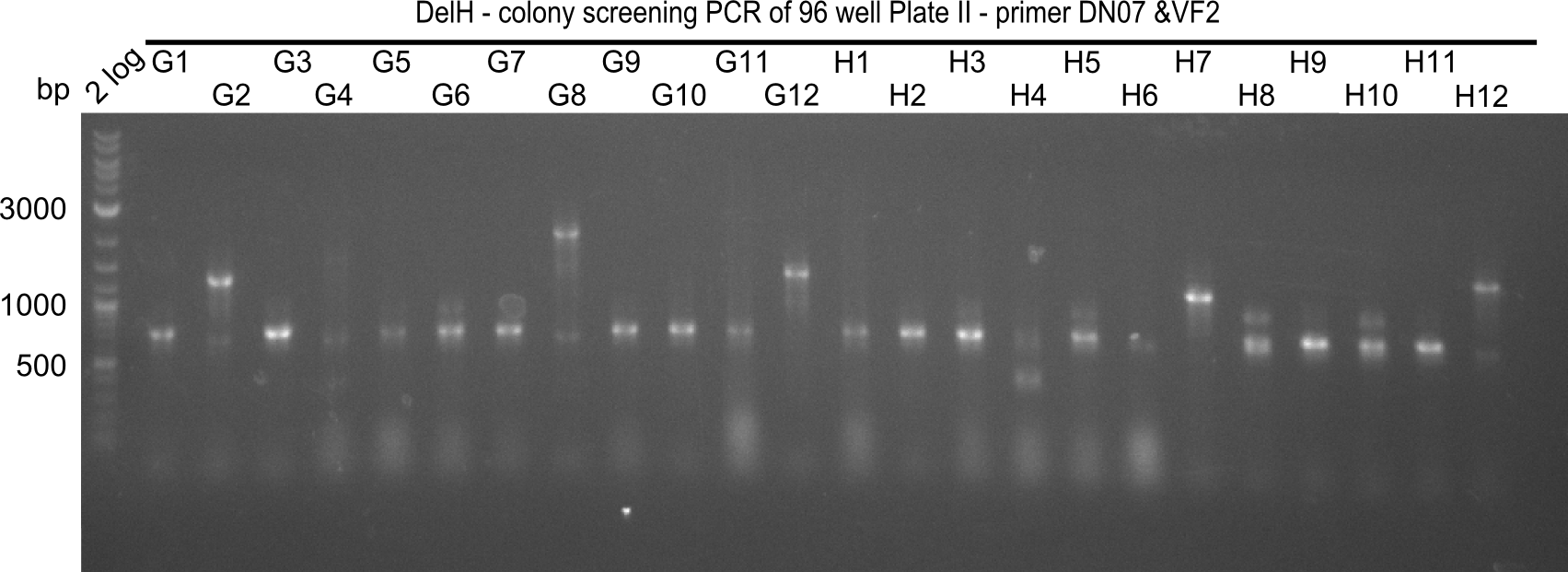
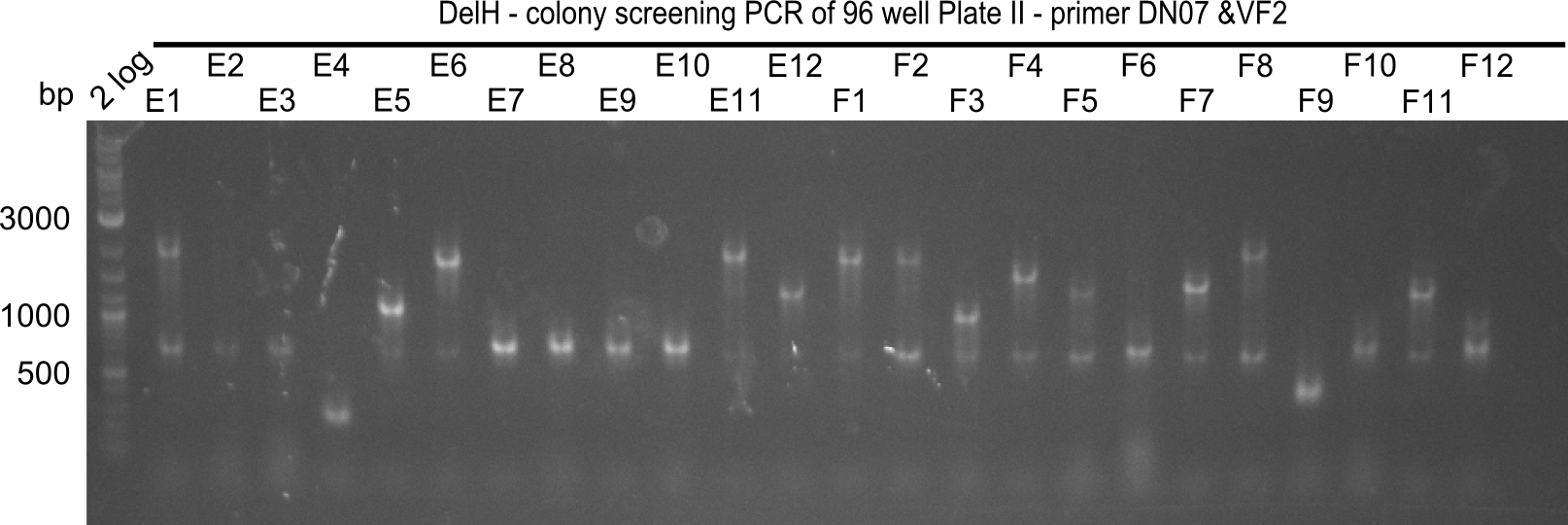
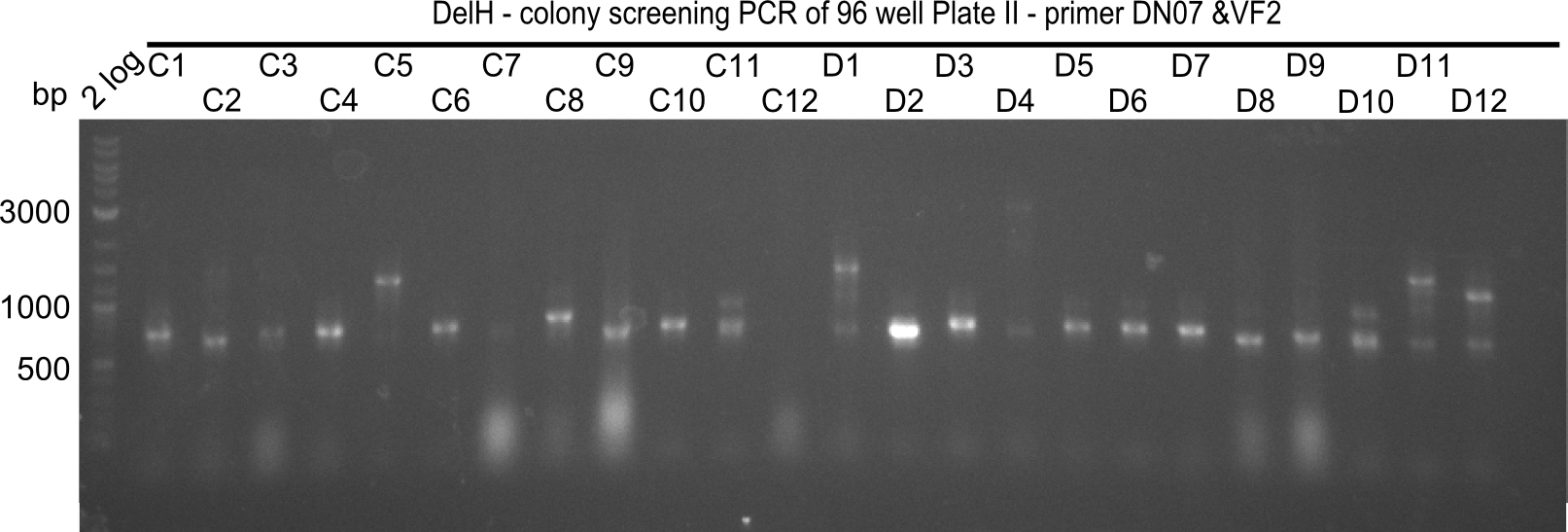
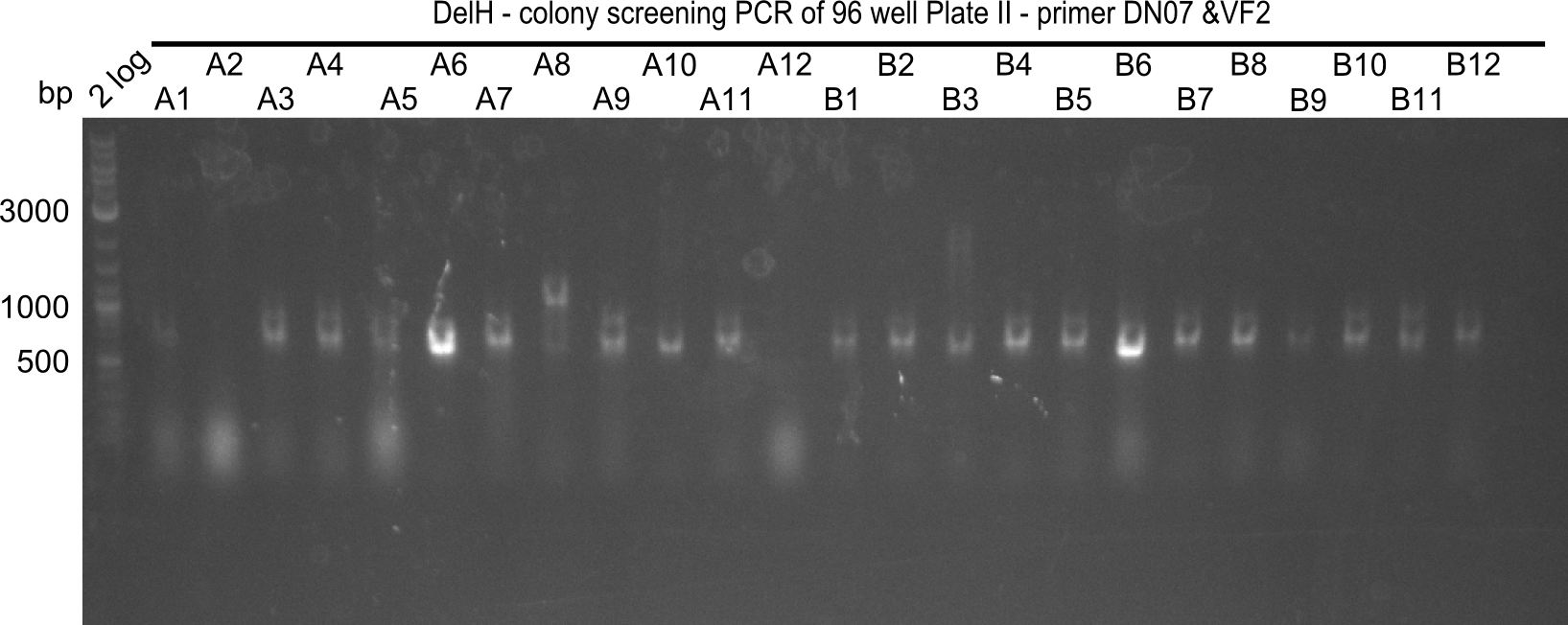
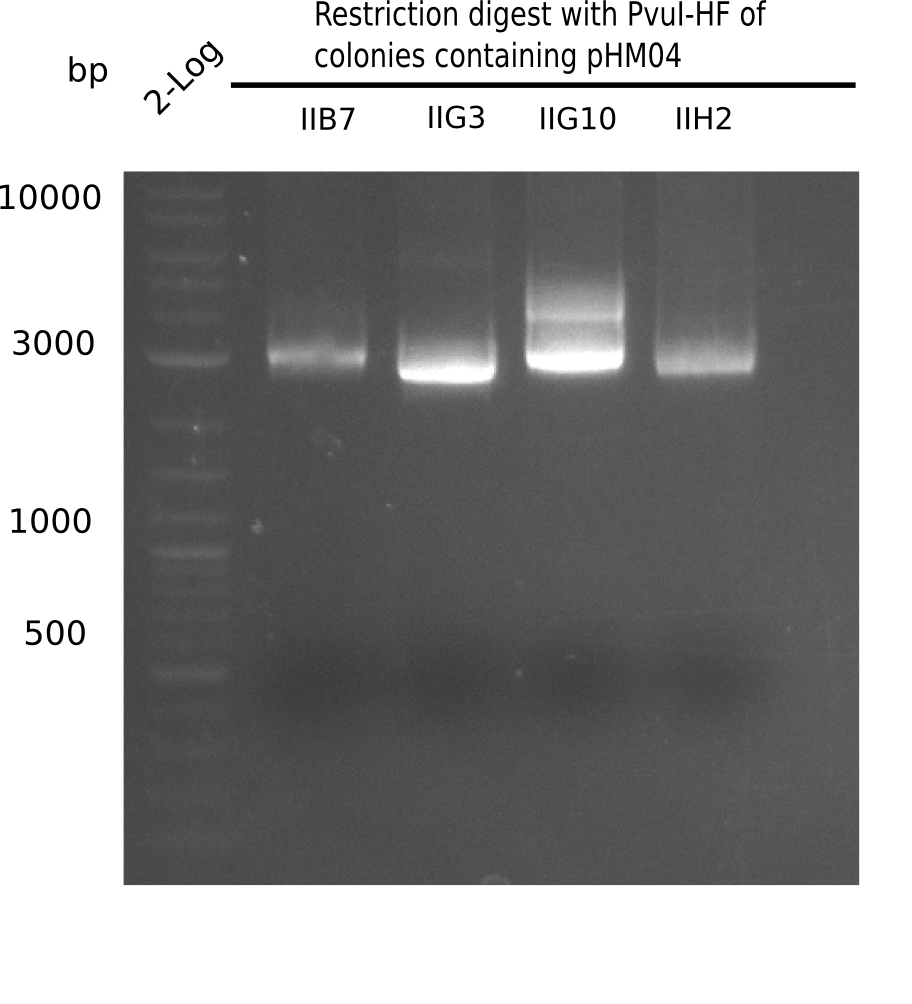
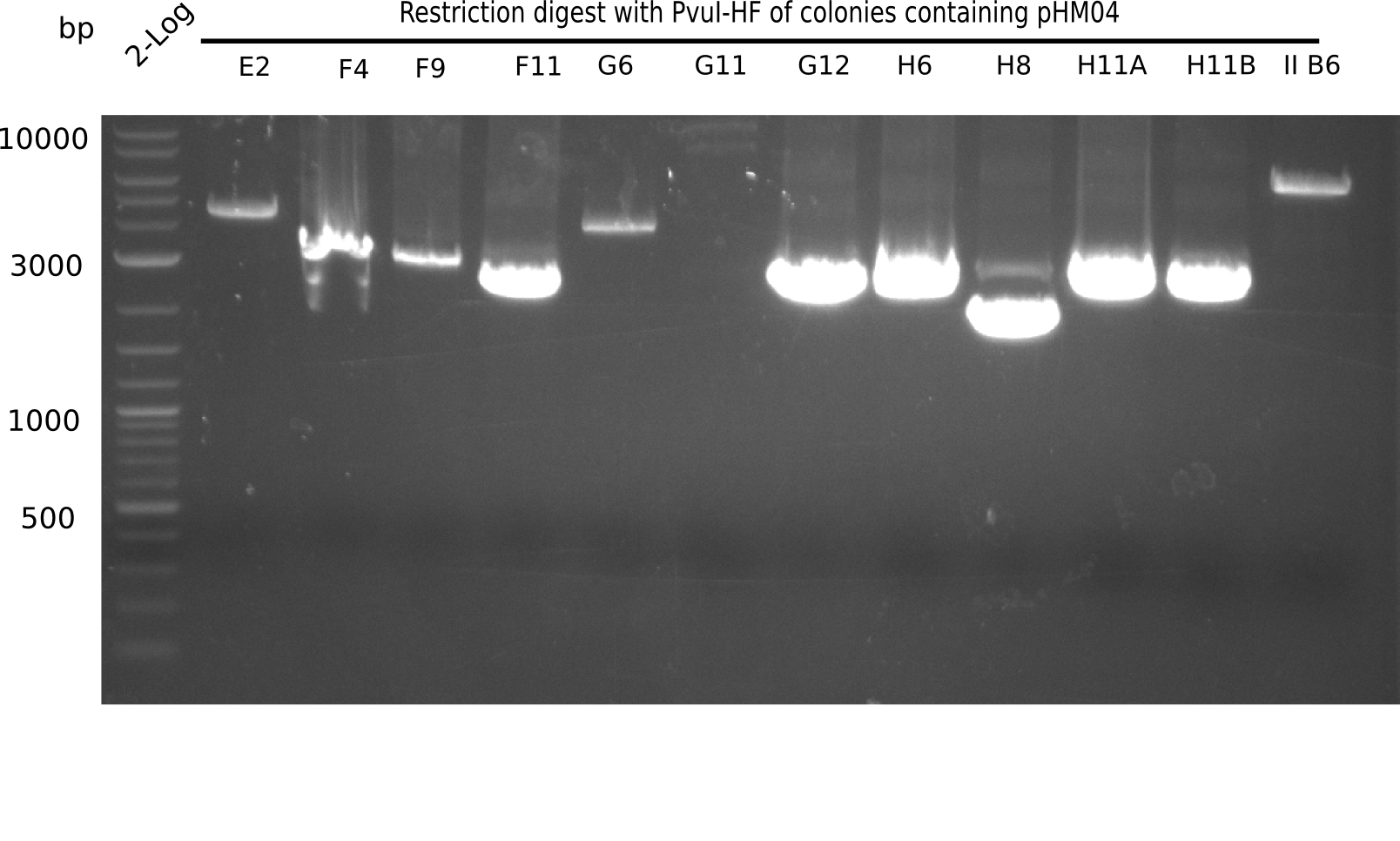
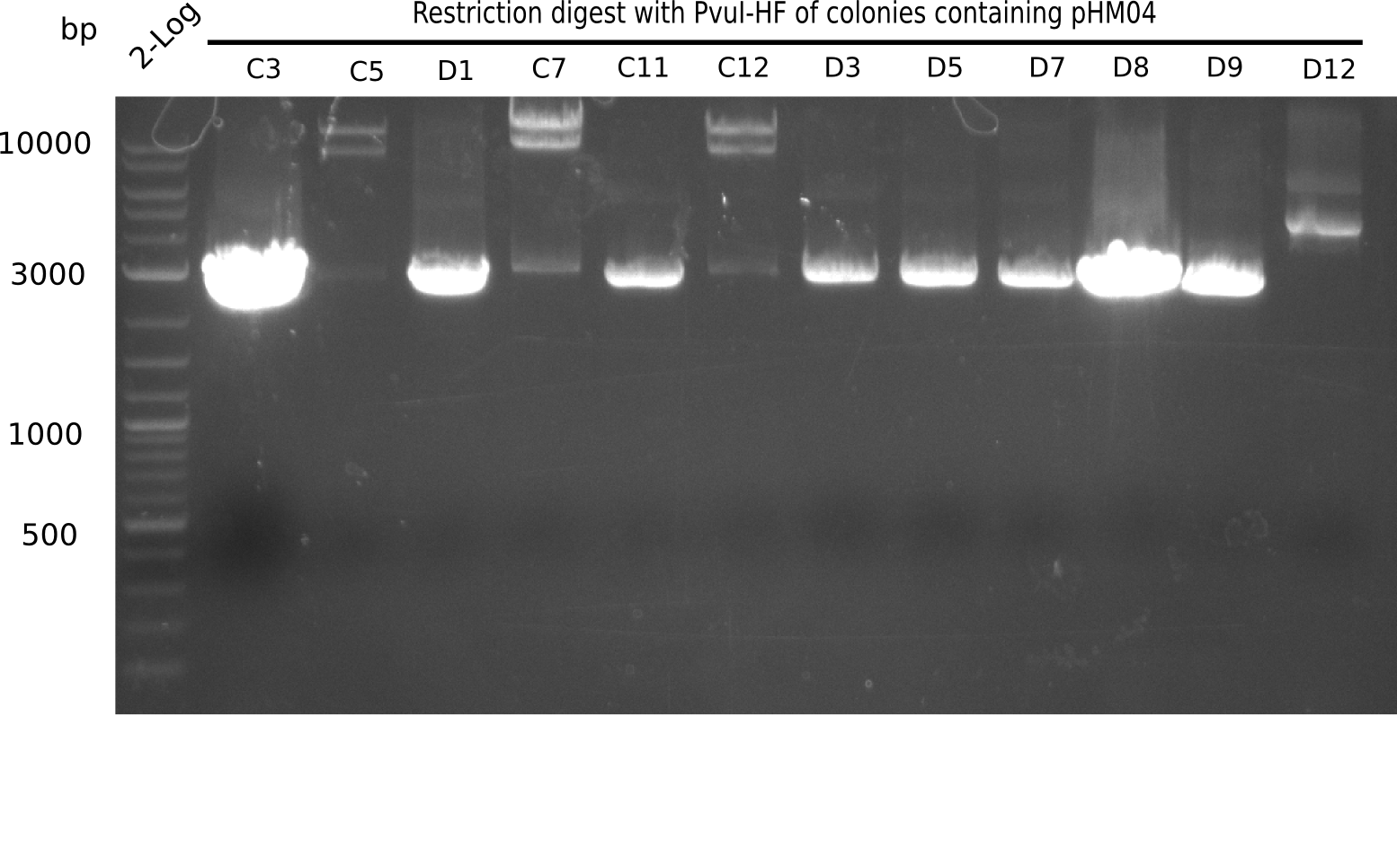
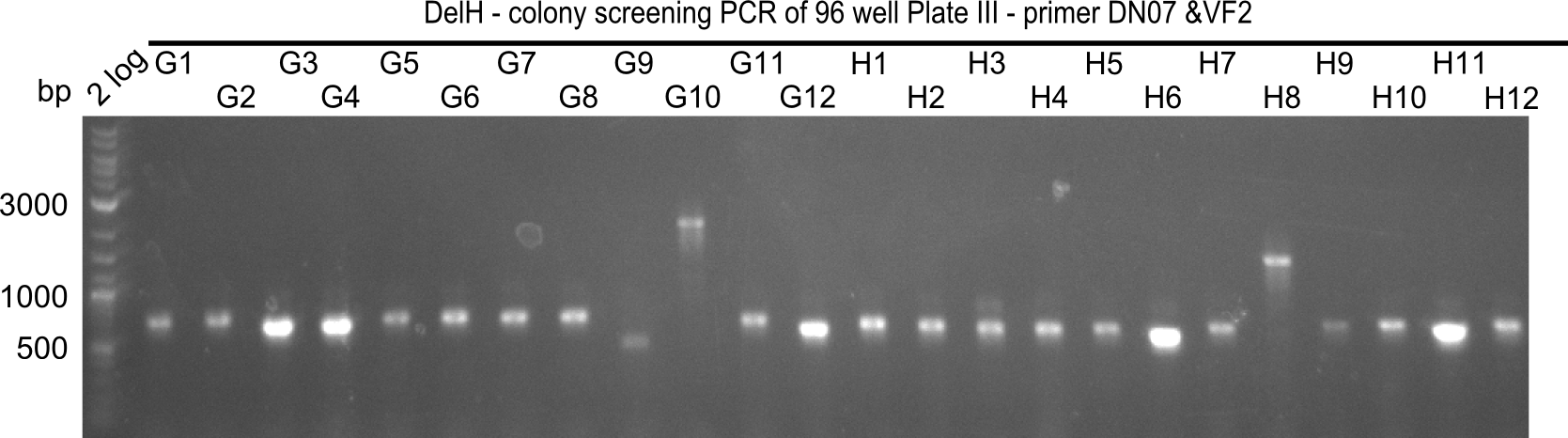
Week 19
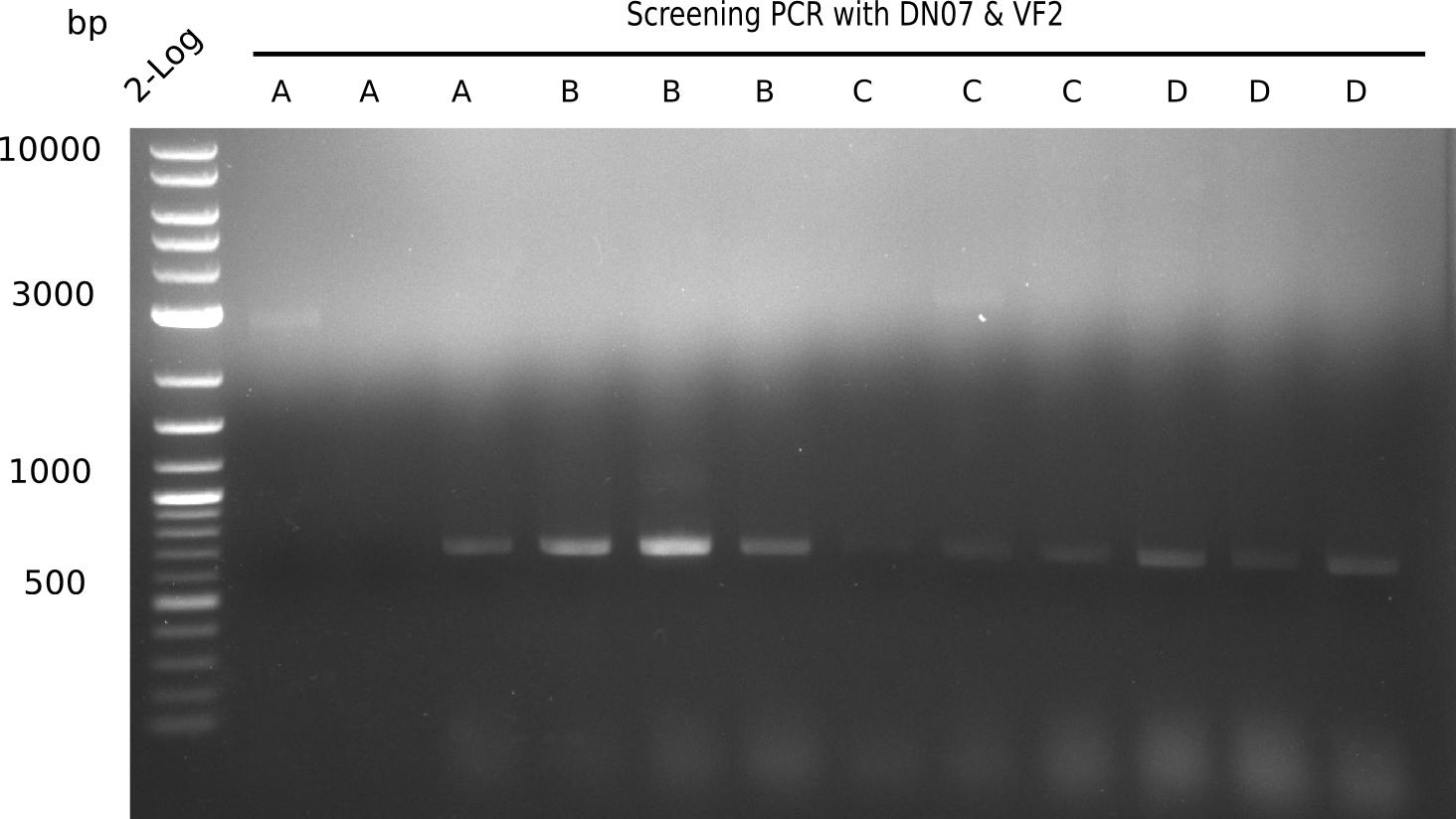
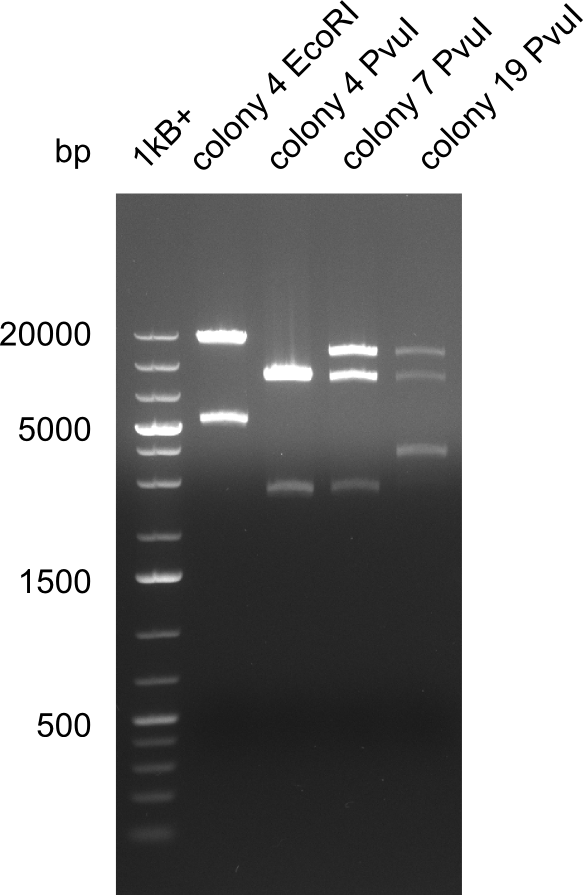
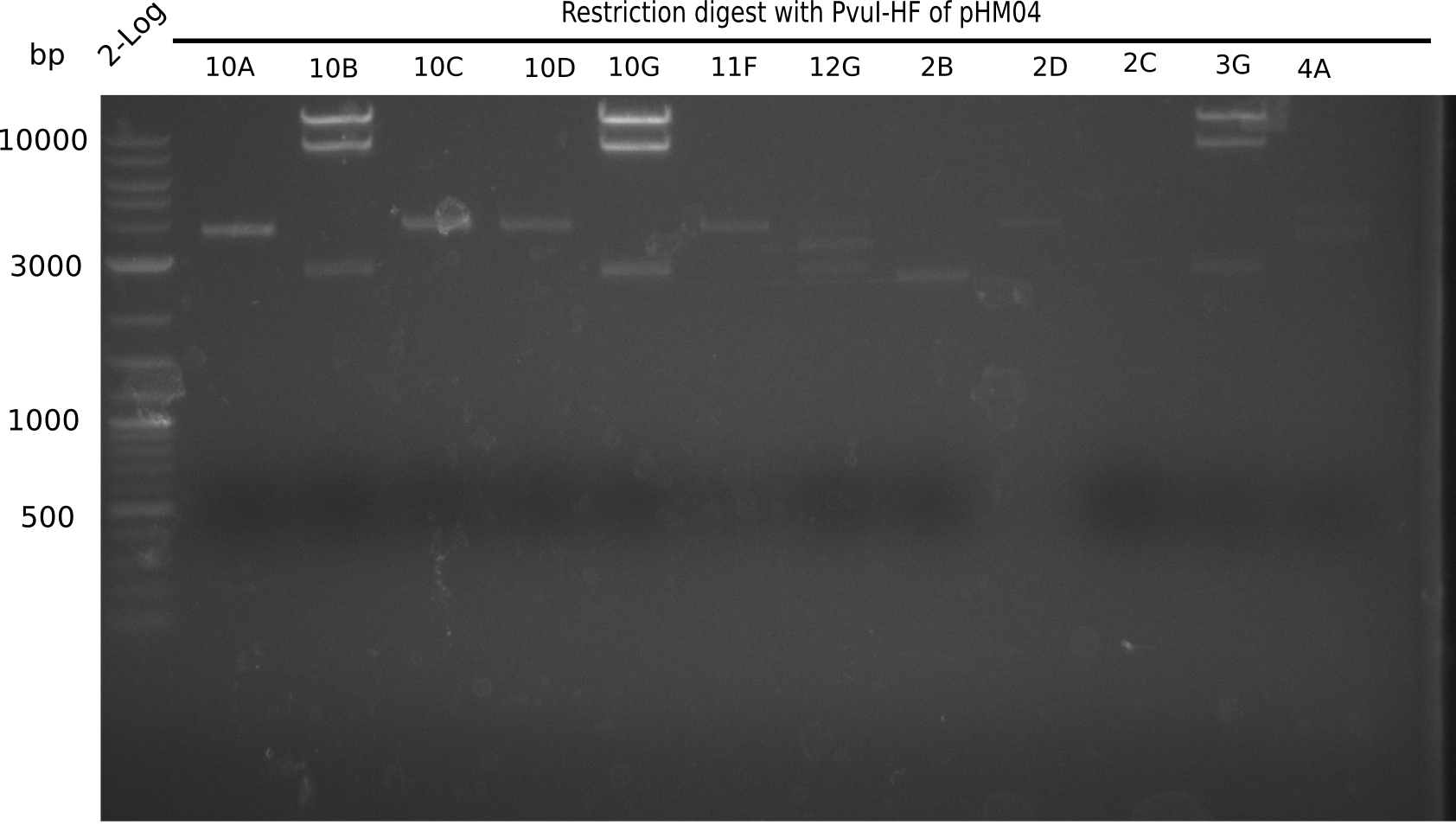
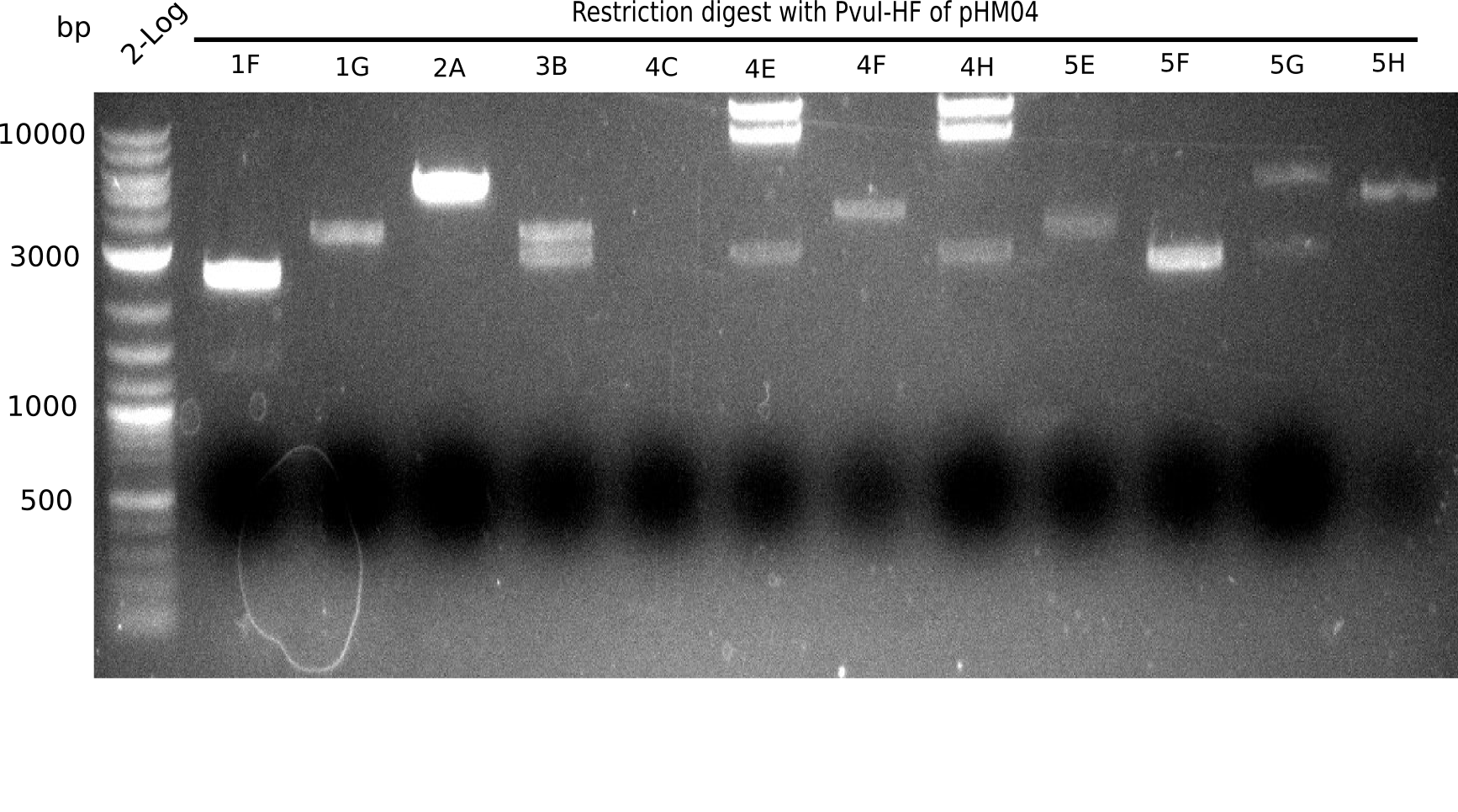
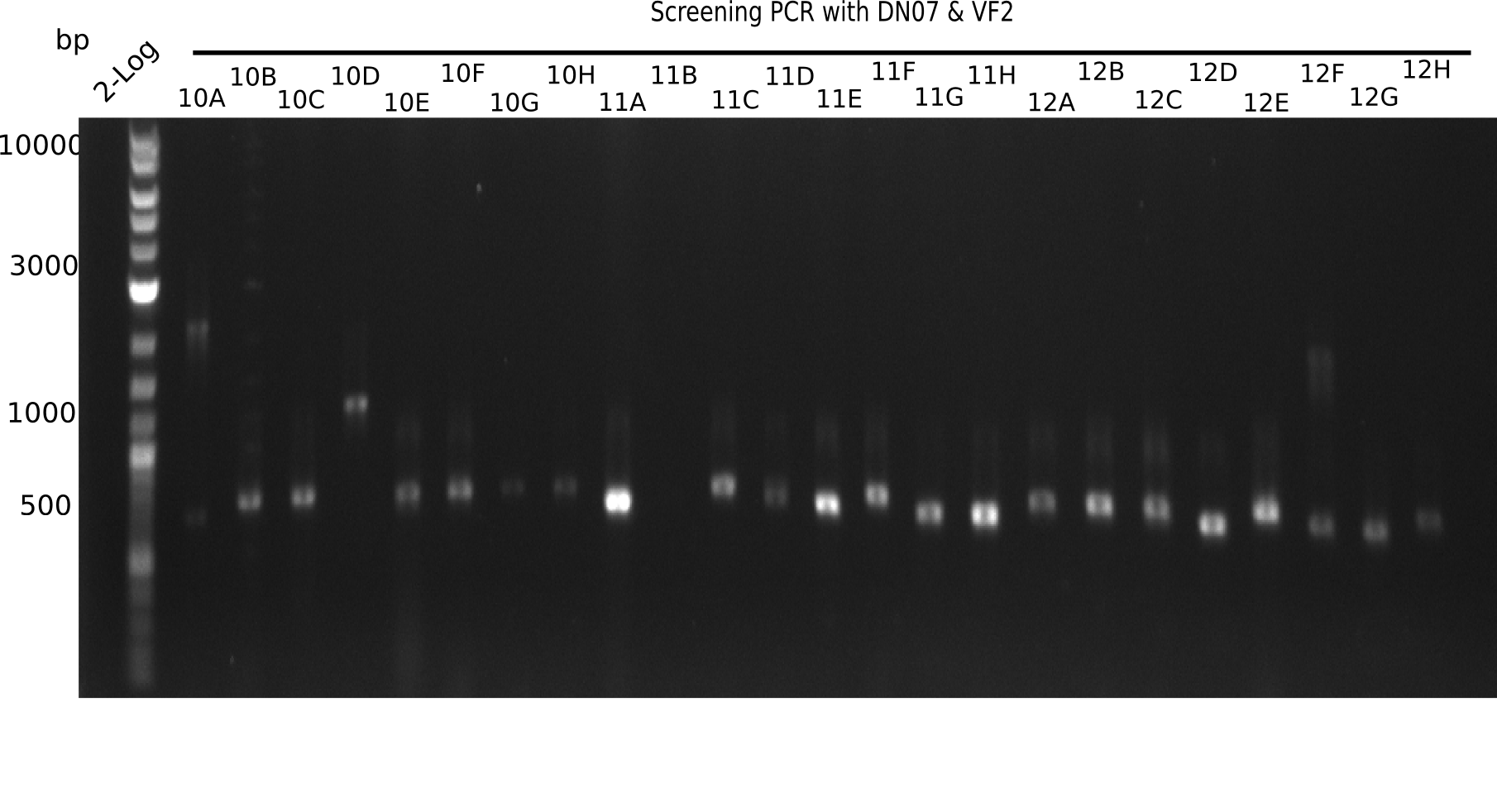
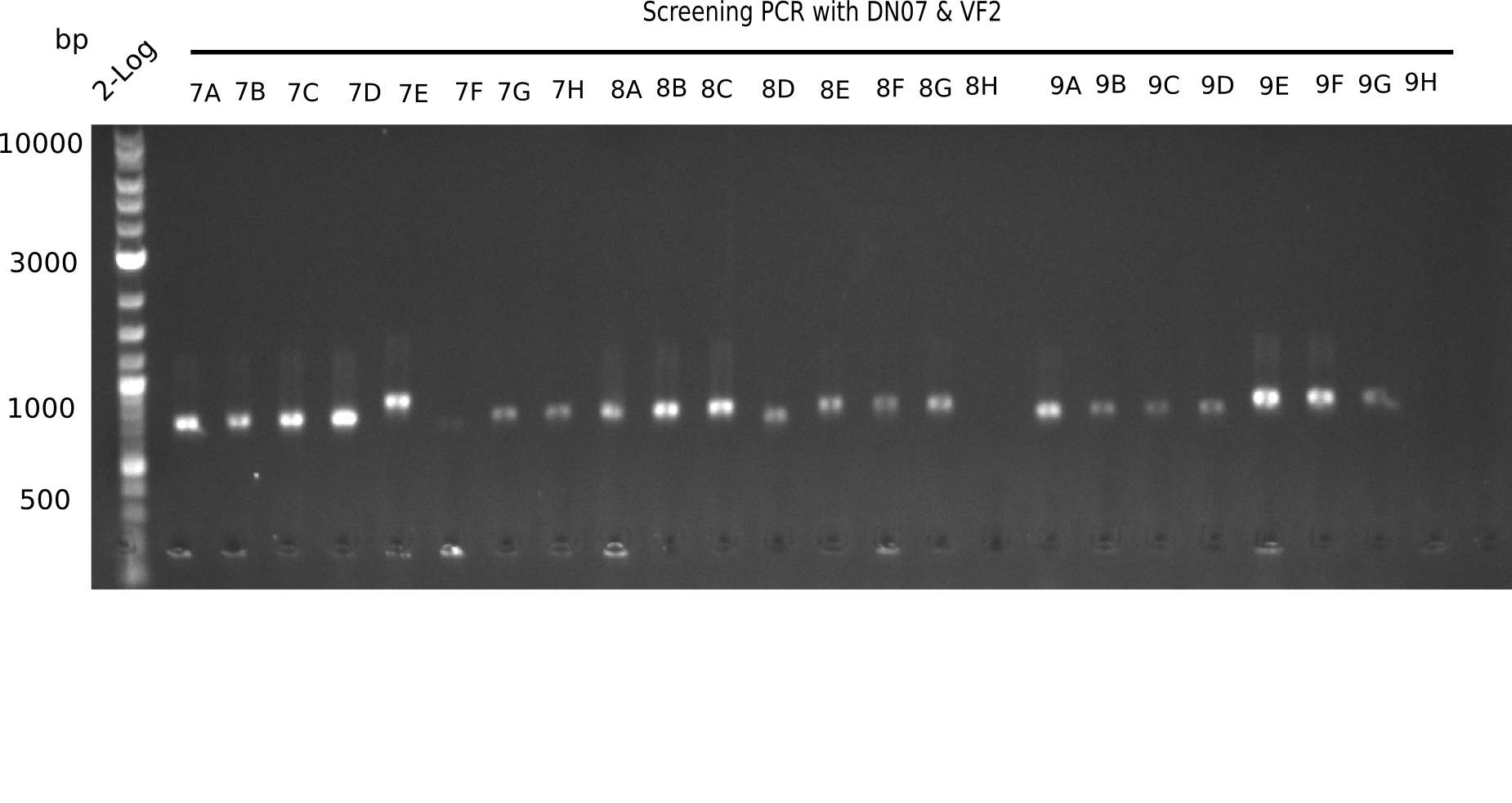
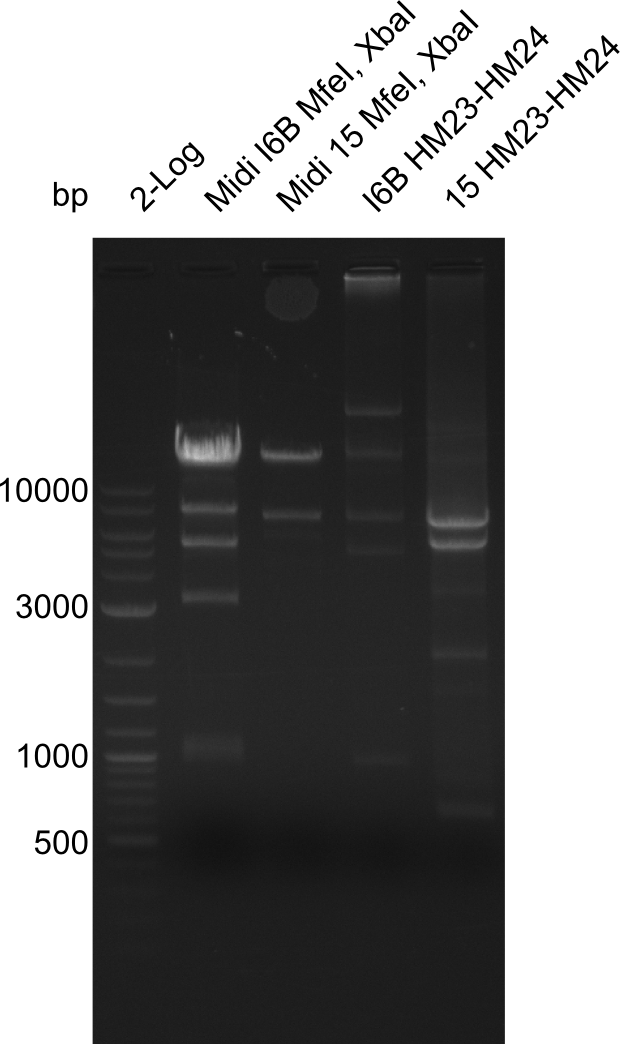
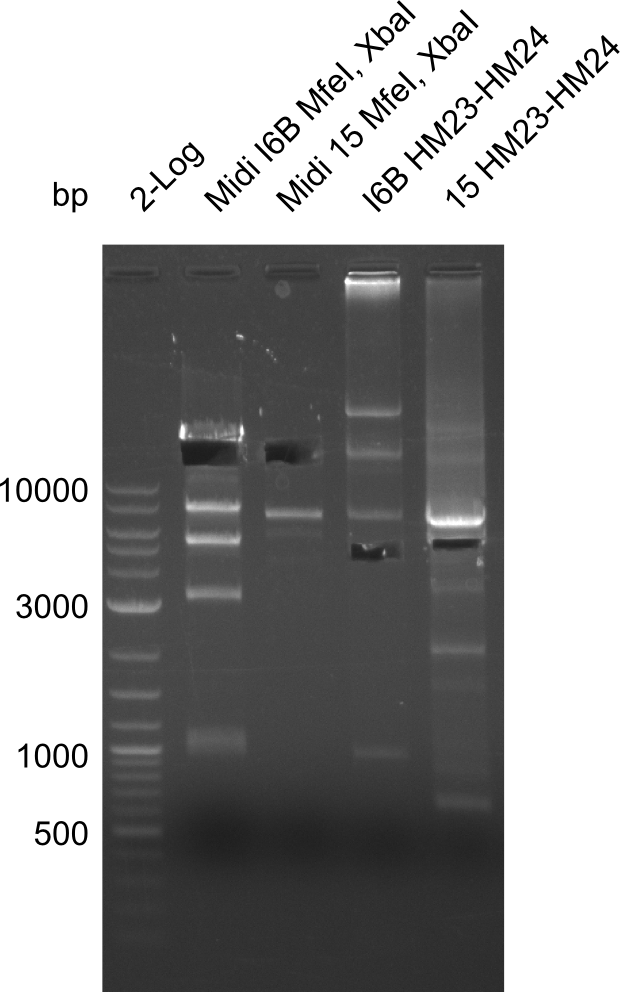
After a successful electroporation, we screen numerous clones by colony-PCR and test restriction digest (PvuI-HF). Positive clones are send for sequencing. The sequencing tells us if the DelH Gibson assembly actually worked. Therefore, we send the midipreped plasmids with the reverse DN07 primer or VF2, covering the start sequence of DelH and the end of the pSB6A1 backbone without mRFP (pHM04). The screening-PCR showed different positive colonies. After performing the restriction digest, some colonies were discarded and the rest was sent in for sequencing. We did not find any positive clone without a truncating mutation at the beginning of DelH (in the primer region).
Week 20
Again, the screening-PCRs resulted in many positive colonies. After the restriction digest, many colonies were kicked out. The remaining minipreped colonies were sent in for sequencing. Yet, we did not identify any positive clone which had no mutation at the beginning of DelH (in the primer region).
Week 21
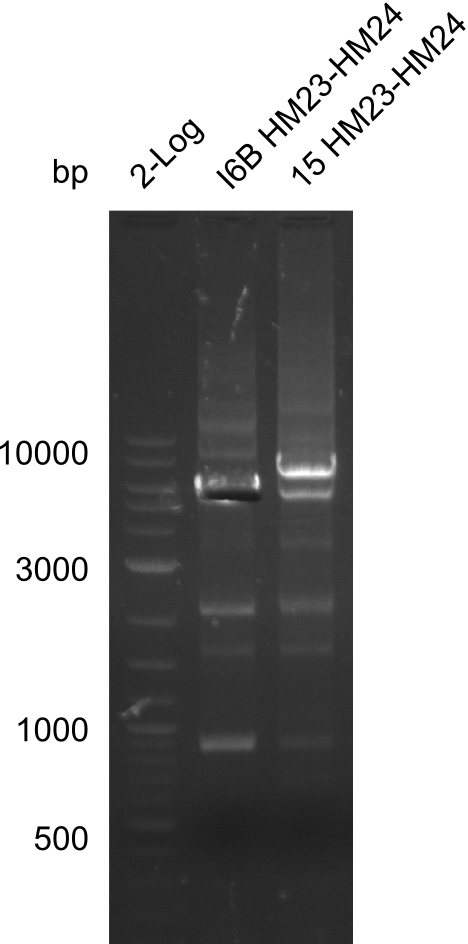
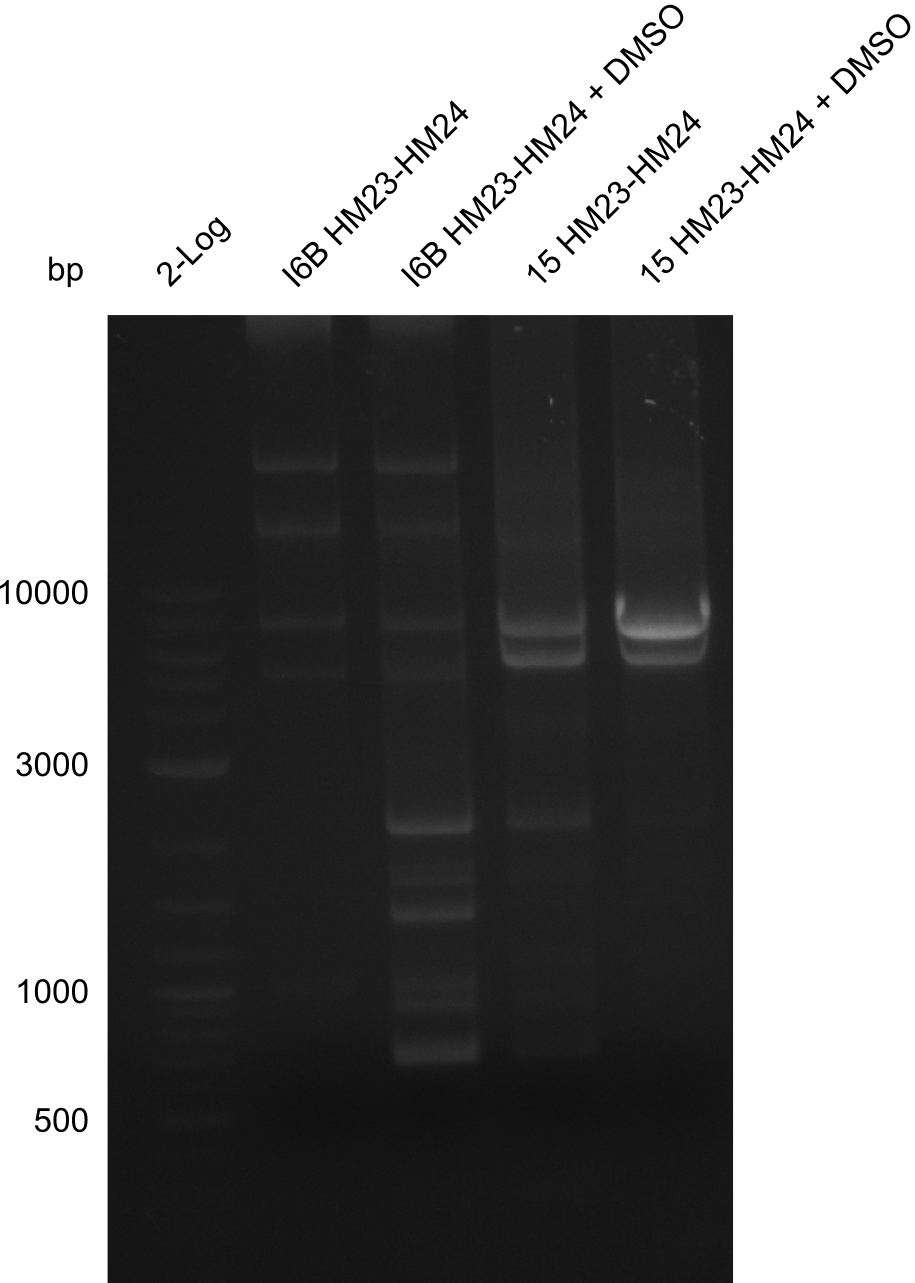
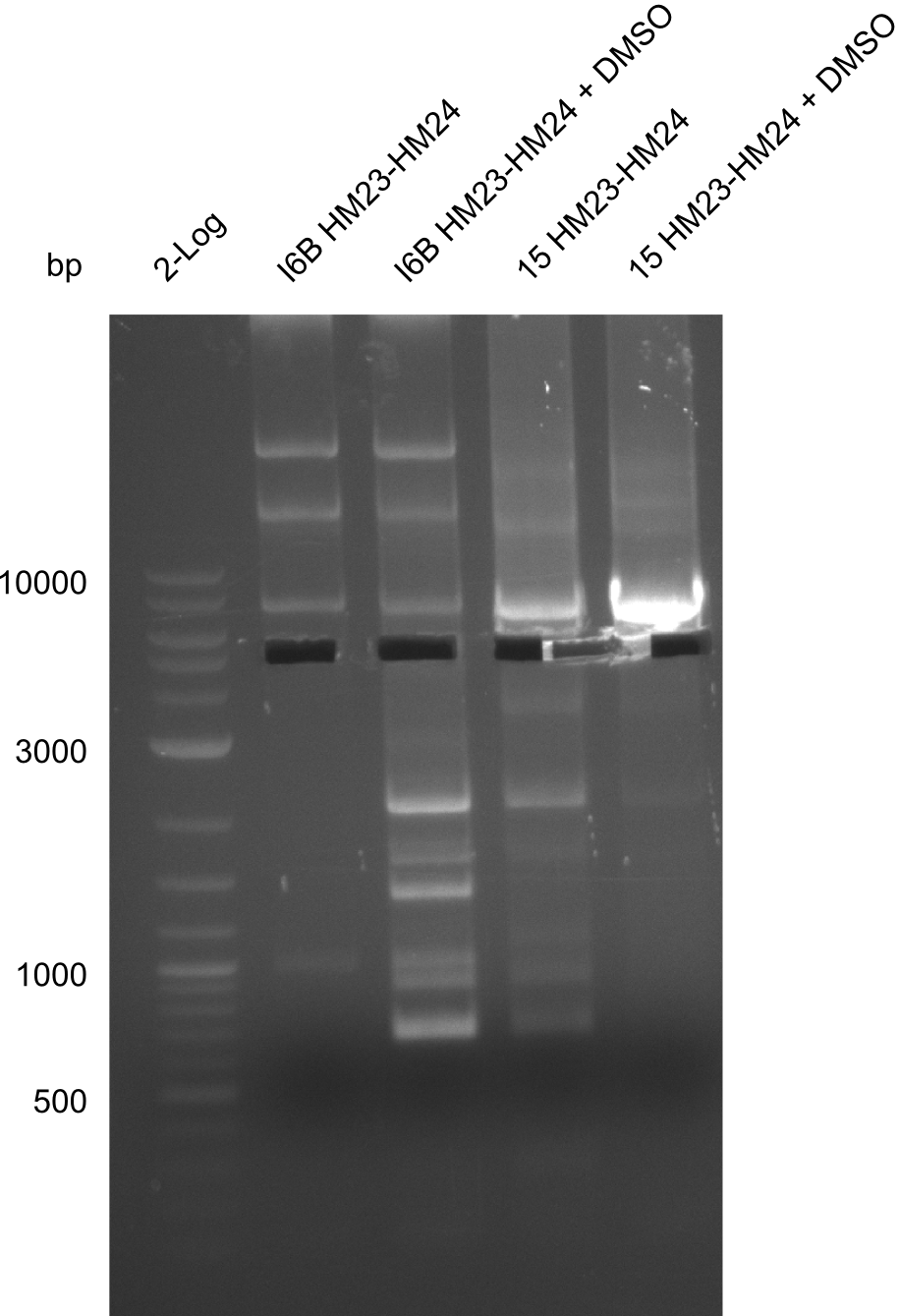
Frustratingly, none of the analyzed clones showed a correct sequence. Probably, DelH by itself is toxic for E.coli and thus, only aberrant clones survive. We suspect the low quality of primers as reason for high number of mutations. Therefore, we will order HPLC purified primers. Additionally, we tried to eliminate the mutations in DelH clones I 6b and 15 by mutagenesis. Herefore, primers were ordered.
Week 22
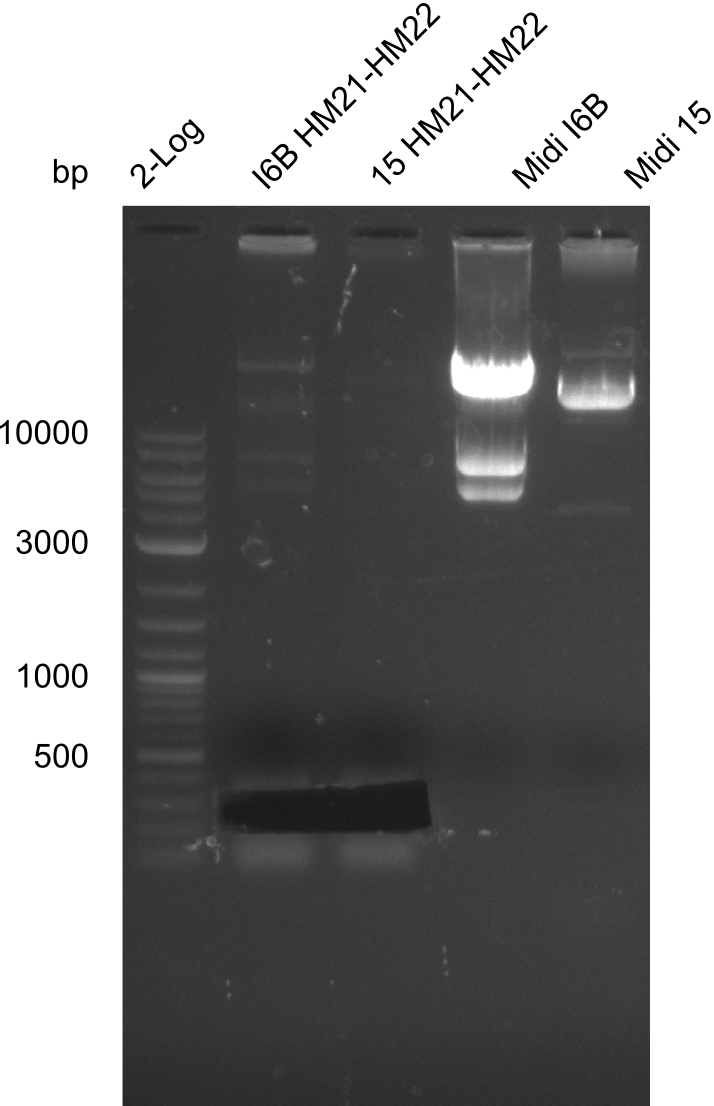
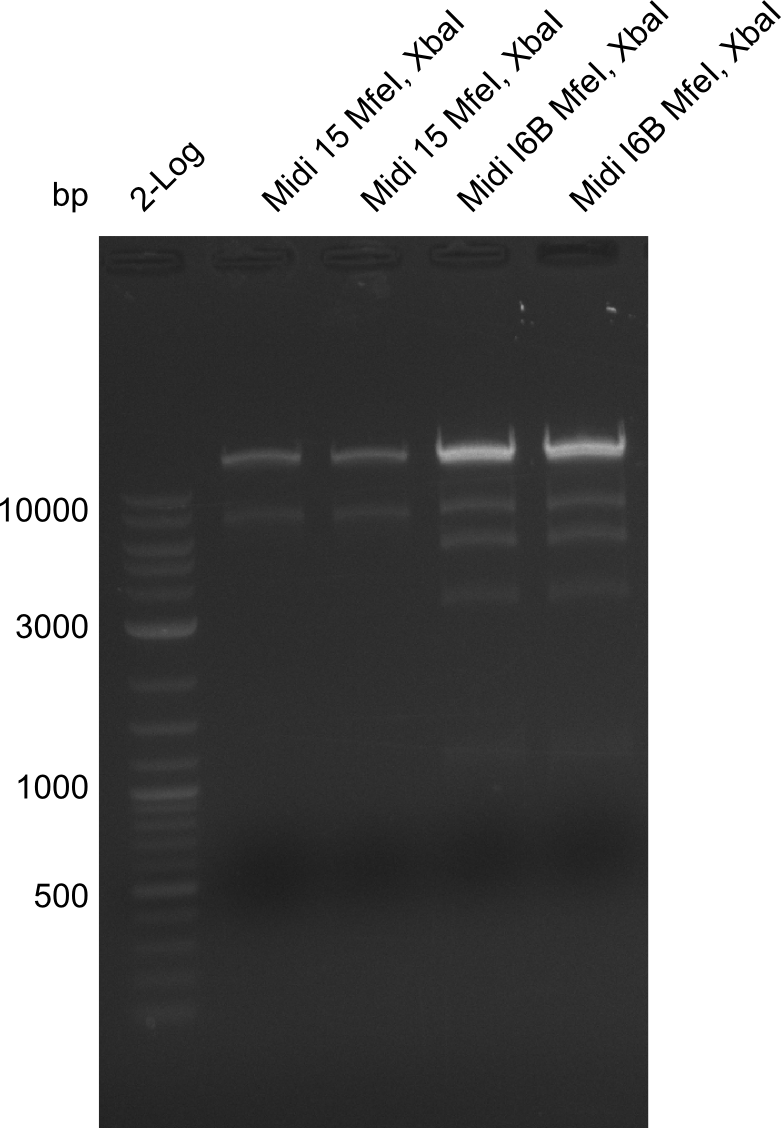
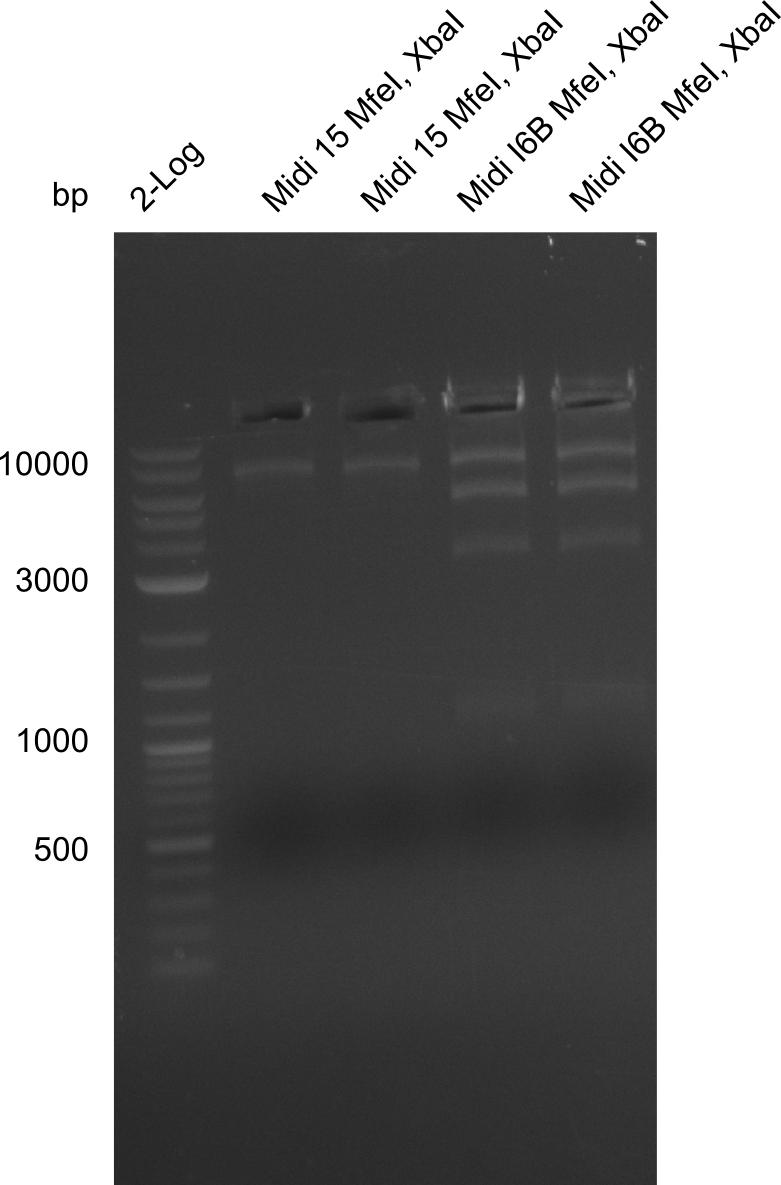
So far, we failed to find a single correct DelH clone. We suspect the separate DelH module to be toxic for the E. coli. Therefore, they select for mutated plasmids. In order to reduce the selection pressure, we used E. coli BL21 DE3, known for increased expression of the lac repressor. This strategy also did not result in any correct clone. One reason might be, that the E. coli BL21 DE3 we obtained actually are BL21 DE3 pLys, which are unfortunately Chloramphenicol resistant due to additional plasmid and thus not useful for screening and amplification of DelH construct. We got new and correct E. coli BL21 DE3 as well as NEB turbo, which significantly overexpress the lac repressor. We however found their lacZ-controlled expression to be very leaky, in contrast to E. coli BL21 DE3. Since the Gibson assembly of DelH using the HPLC purified primers also exclusively resulted in mutated clones, we developed two new cloning strategies to avoid expression of DelH. The first strategy uses a weak promoter and RBS, the second introduces DelH in a ccdB helper construct. Last but not least, we continued working on the mutagenesis approach to eliminate the mutation in clone C5. It was sent for sequencing...
Week 23
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Methods:
Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet. Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet.Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. .
29-04 - 05-05-13
Generation of Plasmid Backbones
Transformation of Biobricks
| Part of the registry | Plate of 2012 | Well | Resistance |
|---|---|---|---|
| pSB4K5 (insert=J04450) | 1 | 5G | Kanamycin |
| pSB6A1 (insert=J04450) | 1 | 1K | Ampicillin |
| lacZ (I732019) in pSB1AK3 | 4 | 12G | Kanamycin, Ampicillin |
| AraC (I0500) in pSB2K3 | 1 | 14N | Kanamycin |
| pSB1C3 (insert=J04450) | 1 | 3A | Chloramphenicol |
- Added 10 µl H2O to each well
- Incubated for 10 min at RT
- Thawed 5x chemical competent E.coli Top10
- 3 µl of plasmid DNA were added
- Incubated for 10 min on ice
- Heat shock for 40 s at 42.2°C
- Incubated for 10 min on ice
- Added 500 µl LB Medium
- Incubated at 37°C for 40 min
- Centrifuged 120 at 5,000 rpm, supernatant discarded
- Pellet resuspended in remaining medium
- Plated on plates with the corresponding antibiotics (as shown in the table above, section: resistances)
- Incubated for 2 days at RT
- One colony was picked from each plate and incubated over night at 37°C in LB medium with the antibiotic listed above
Result
All 5 parts from the Registry Distribution 2012 were successfully transformed in E.coli Top10 except for the one containing the AraC promotor.
Amplification of DelH Fragments
Design of Primers
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| DN01:DelH_f1_PacI_fw | 03-05-2013 | Amplification of DelH F1, with RBS and adding PacI restriction site | TTTT TTAATTAA TCACACAGGAAAGTACTAG ATGGACCGTGGCCGCCTGC GCCAAATCG |
| DN02:DelH_f1_SalI_rev | 03-05-2013 | Amplification of DelH F1 until SalI restriction site | TTTT GTCGACCAACACCTGTGCCTGC |
| DN03:DelH_f2_SalI_fw | 03-05-2013 | Amplification of DelH F2 starting at SalI restriction site | TTTT GTCGACTGGATGGAGCCTGGTGAAAG |
| DN04:DelH_f2_KpnI_rev | 03-05-2013 | Amplification of DelH F2, adding KpnII restriction site | TTTT GGTACC TCAGTCCAGCGCGTACTCCAG |
| DN05:AraCbb_KpnI_fw | 03-05-2013 | Amplification of backbone for DelH (pSB6A1-AraC-lacZ), adding KpnI site | TTTT GGTACC AAAGAGGAGAAATACTAGATGACCATG |
| DN08:AraCbb_PacI_rev | 03-05-2013 | Amplification of backbone for DelH (pSB6A1-AraC-lacZ), adding PacI site | TTTT TTAATTAA GCTAGCCCAAAAAAACGGGTATG |
Amplification of 2
06-05 - 12-05-13
Amplification of BioBricks
Miniprep of BioBricks
- Miniprep was performed using Qiagen kit and eluted in 35 µl ddH2O.
- From 500 µl of liquid culture, a glycerol stock was prepared as described in Preparing glycerol stocks.
Result
- DNA concentration was determined using nanovue.
| Sample | Concentration [ng/µl] |
|---|---|
| pSB4K5 | 21 |
| pSB6A1 | 47 |
| lacZ | 45 |
| pSB1C3 | 23 |
Generation of Plasmid Backbones
Restriction Digest of BioBrick pSB6A1
| Component | Amount [µl] |
|---|---|
| PSB6A1 DNA | 32.5 |
| BSA (10x) | 5 |
| NEB2 buffer (10x) | 5 |
| Enzymes (EcoRI-HF and PstI) | 1.5 each |
| ddH2O | 4.5 |
Result
The fragments of plasmids pSB4K5 and pSB1C3 show the expected pattern.
- => BioBrick backbone was cut and gel isolated.
Generation of LacZ Fragment
Restriction Digest of BioBrick pSB1AK3 Conditions A
| Component | Amount [µl] |
|---|---|
| pSB1AK3 DNA | 32.5 |
| BSA (10x) | 5 |
| NEB3 buffer (10x) | 5 |
| Enzymes (XbaI and PstI) | 1.5 each |
| ddH2O | 4.5 |
- Incubation for 1 h at 37°C
Result
- The restriction mix was loaded onto a 1% agarose gel using 6 µl gene ruler marker.
The fragments show the expected pattern.
- => LacZ insert were cut and gel isolated.
Restriction Digest of BioBrick K173004(lacZ) Conditions B
| Component | Amount [µl] |
|---|---|
| pSB1AK3 DNA | 8.3 |
| BSA (10x) | 5 |
| NEB4 buffer (10x) | 5 |
| Enzymes (XbaI & SalI-HF) | 1.5 each |
| ddH2O | 28.7 |
- Purification was performed with Qiagen Nucleotide removal Kit and eluted in 38.5 µl ddH2O
Result
2.5 µl were measured with the Nanovue. The measurement proved the failure of miniprep.
Restriction Digest of BioBrick K173004(lacZ) Conditions C
| Component | Amount [µl] |
|---|---|
| pSB1AK3 DNA | 32.5 |
| BSA (10x) | 5 |
| NEB 3 buffer (10x) | 5 |
| Enzyme (PstI) | 1.5 |
| ddH2O | - |
Result
- =>The band was too high on gel and discarded, because we cannot be sure if it really is lacZ or the backbone of the lacZ plasmid.
Generation of AraC Fragment
Amplification of BioBricks containing I13453 and K206000
- The transformation was performed as described on 03-05 and incubated ON at 37°.
Amplification of DelH F1
PCR Conditions F1.W2.A
| Reagent | DelH F1 | DelH F1 |
|---|---|---|
| Expected length [Kb] | 10 | 10 |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans DNA |
| Primer 100 µM fw | 0.5 µl DelH_f1_PacI_fw | 0.5 µl DelH_f1_PacI_fw |
| Primer 100 µM rev | 0.5 µl DelH_f1_SalI_rev | 0.5 µl DelH_f1_SalI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| ddH2O | 23 µl | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 |
| 66(↓0.5°C) | 5 | |
| 72 | 3:40 min | |
| 14 | 98 | 1 |
| 62 | 5 | |
| 72 | 3:40 min | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 10 Kb
Different unspecific bands were observed.
- => Further optimize PCR conditions.
PCR Conditions F1.W2.B
| Reagent | DelH-fragment1 | DelH-fragment1 |
|---|---|---|
| Expected length [Kb] | 10 | 10 |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans DNA |
| Primer 100 µM fw | 0.5 µl DelH_f1_PacI_fw | 0.5 µl DelH_f1_PacI_fw |
| Primer 100 µM rev | 0.5 µl DelH_f1_SalI_rev | 0.5 µl DelH_f1_SalI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| ddH2O | 23 µl | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 2:45 min | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
PCRs showed only unspecific bands.
- => Further optimize PCR conditions.
PCR Conditions F1.W2.C
| Reagent | DelH F1 | DelH F1 | DelH F1 | DelH F1 |
|---|---|---|---|---|
| Expected length [Kb] | 10 | 10 | 10 | 10 |
| Template | 1 µl D. acidovorans normal | 1 µl D. acidovorans normal | 1 µl D. acidovorans 1:25 | 1 µl D. acidovorans 1:25 |
| DelH_f1_PacI_fw | 0.5 µl normal | 0.5 µl 1:10 | 0.5 µl normal | 0.5 µl 1:10 |
| DelH_f1_SalI_rev | 0.5 µl normal | 0.5 µl 1:10 | 0.5 µl normal | 0.5 µl 1:10 |
| Phusion Master Mix (2x) | 25 µl | 25 µl | 25 µl | 25 µl |
| DMSO | - | - | 2.5 µl | 2.5 µl |
| ddH2O | 23 µl | 23 µl | 20.5 µl | 20.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 |
| 62 | 5 | |
| 72 | 2:45 min | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
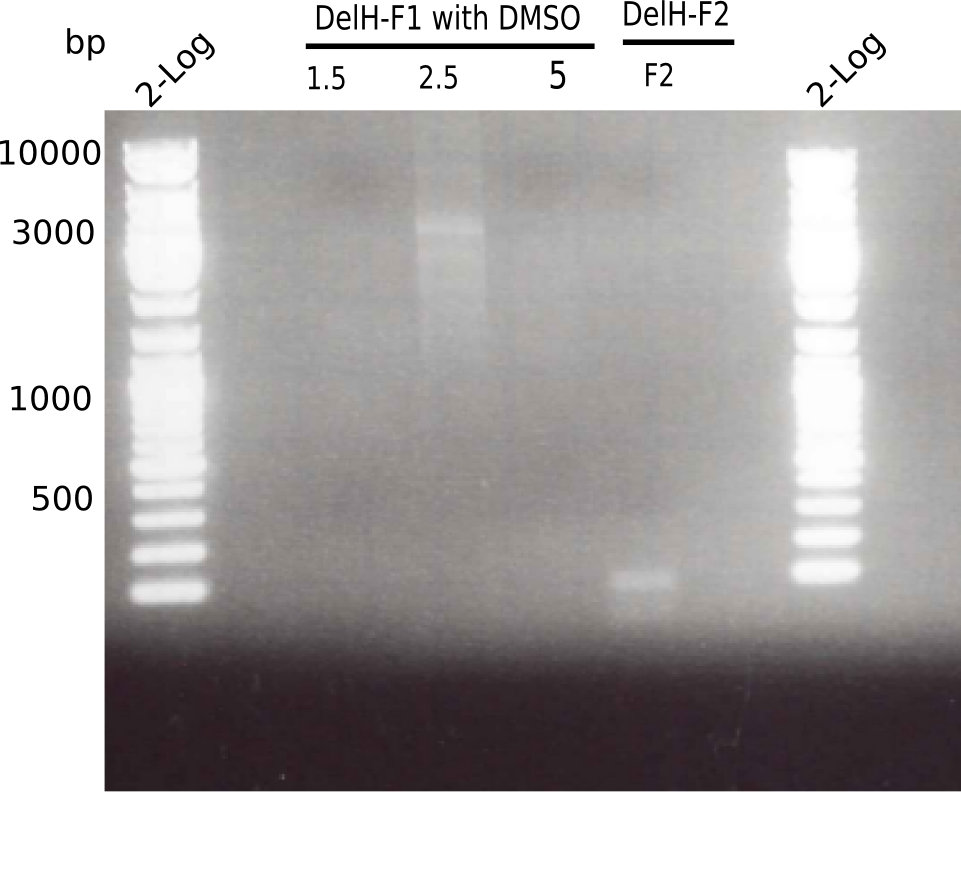
PCR with DMSO and 1:10 diluted primer looked best.
- => Gel extraction performed.
Amplification of DelH F2
PCR Conditions F2.W2.A
| Reagent | DelH F2 | DelH F2 |
|---|---|---|
| Expected length [Kb] | 8 | 8 |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans DNA |
| Primer 100 µM fw | 0.5 µl DelH_f2_SalI_fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 100 µM rev | 0.5 µl DelH_f2_KpnI_rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| ddH2O | 23 µl | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 |
| 66(↓0.5°C) | 5 | |
| 72 | 3:40 min | |
| 14 | 98 | 1 s |
| 62 | 5 s | |
| 72 | 3:40 min | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 8 Kb
Different unspecific bands were observed.
- => Further optimize PCR conditions.
PCR Conditions F2.W2.B
| Reagent | DelH F2 | DelH F2 |
|---|---|---|
| Expected length [Kb] | 8 | 8 |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans DNA |
| Primer 100 µM fw | 0.5 µl DelH_f2_SalI_fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 100 µM rev | 0.5 µl DelH_f2_KpnI_rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| ddH2O | 23 µl | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 s |
| 72 | 2:45 min | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Results
Expected band: 8 Kb
Gel shows the expected specific band at 8 Kb.
- => DelH F2 was thus successfully amplified from the glycerol stock.
Amplification of 1
13-05 - 19-05-13
Amplification of DelH F1
Gel Purification
45 µl of DelH F1 PCR (08-05) product were run on a 0,8% agarose gel (1 h with 135V). Gel extraction was performed using gel extraction kit of Qiagen, and diluted in 25 µl ddH2O.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| DelH F1 | 7 |
- => Because the concentration is so low, a re-PCR of the PCR product (08-05) is going to be done.
re-PCR Conditions F1.W3.A
| Reagent | DelH F1 |
|---|---|
| Expected length [Kb] | 10 |
| Template | 1 µl 1:10 gel purified F1 |
| Primer 10 µM fw | 0.5 µl DelH_f1_PacI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f1_SalI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 2.5 µl |
| ddH2O | 20.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:30 min | |
| 72 | 3:30 min | |
| 1 | 4 | inf |
Result
Expected length: 10 Kb
Different unspecific bands were observed.
- => Test digest to confirm identity.
Test Restriction Digest
| PCR product of DelH-F1 | BSA | NEBuffer 4 | Enzymes | ddH2O | Total amount |
|---|---|---|---|---|---|
| 18 µl | 5 µl | 5 µl | 2x 1.5 µl SalI-HF & PacI | 19 µl | 50 µl |
Afterwards, purification with the nucleotide removal kit was performed, but due to using wrong column, there was no product left.
- => Repeat amplification of DelH F1.
PCR Conditions F1.W3.B
| Reagent | DelH F1 | DelH F1 | DelH F1 |
|---|---|---|---|
| Expected length [Kb] | 10 | 10 | 10 |
| Template | Picked colony | Picked colony | Picked colony |
| Primer 10 µM fw | 0.5 µl DelH_f1_PacI_fw | 0.5 µl DelH_f1_PacI_fw | 0.5 µl DelH_f1_PacI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f1_SalI_rev | 0.5 µl DelH_f1_SalI_rev | 0.5 µl DelH_f1_SalI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl | 25 µl |
| DMSO | 1.5 µl | 2.5 µl | 5 µl |
| ddH2O | 21.5 µl | 20.5 µl | 18 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 3:30 min | |
| 1 | 72 | 3:30 min |
| 1 | 4 | inf |
Result
Expected length: 10 Kb
There is only an unspecific band at the second PCR of F1 with 2.5 µl DMSO.
- => Repeat using DMSO and altered PCR program.
PCR Conditions F1.W3.C
| Reagent | DelH F1 | DelH F1 |
|---|---|---|
| Expected length [Kb] | 10 | 10 |
| Template | 1 µl glycerol stock | 1 µl glycerol stock |
| Primer 10 µM fw | 0.5 µl DelH_f1_PacI_fw | 0.5 µl DelH_f1_PacI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f1_SalI_rev | 0.5 µl DelH_f1_SalI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| DMSO | 2.5 µl | 2.5 µl |
| ddH2O | 20.5 µl | 20.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 3:30 min | |
| 1 | 72 | 3:30 min |
| 1 | 4 | inf |
Result
Expected length: 10 Kb
There was no band visible.
- => Repeat using DMSO and altered PCR program.
Amplification of DelH F2
Gel Purification
45 µl of DelH F1 PCR (07-05) product were run on a 0,8% agarose gel (1 h with 135V). Gel extraction was performed using gel extraction kit of Qiagen, and diluted in 25 µl ddH2O.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| DelH F2 | 6 |
- => Because the concentration is so low, a re-PCR of the PCR product (07-05) is going to be done.
Re-PCR Conditions F2.W3.A
| Reagent | DelH-fragment2 |
|---|---|
| Expected length [Kb] | 8 |
| Template | 1 µl 1:10 gel purified F2 |
| Primer 10 µM fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 2.5 µl |
| ddH2O | 20.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 3:30 min | |
| 72 | 3:30 min | |
| 1 | 4 | inf |
Result
Expected length: 8 Kb
Specific band was observed.
- => Test digest to confirm identity.
Test Restriction Digest
| PCR product of DelH-F1 | BSA | NEBuffer 4 | Enzymes | ddH2O | Total amount |
|---|---|---|---|---|---|
| 18 µl | 5 µl | 5 µl | 2x 1.5 µl SalI-HF & KpnI-HF | 19 µl | 50 µl |
Afterwards, a purification with the nucleotide removal kit was performed, but due to using wrong column, there was no product left.
- => Repeat amplification of DelH F2.
PCR Conditions F2.W3.B
| Reagent | DelH F2 |
|---|---|
| Expected length [Kb] | 8 |
| Template | Picked colony |
| Primer 10 µM fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 0 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:45 min | |
| 1 | 72 | 2:45 min |
| 1 | 4 | inf |
Result
Expected length: 8 Kb
There is no band visible.
- => Repeat using altered PCR program.
PCR Conditions F2.W3.B
| Reagent | DelH F2 |
|---|---|
| Expected length [Kb] | 8 |
| Template | 1 µl glycerol stock |
| Primer 10 µM fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 0 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:45 min | |
| 1 | 72 | 2:45 min |
| 1 | 4 | inf |
Result
Expected length: 8 Kb
There was a specific band at 8 Kb
- => Fragment was cut and gel extracted.
Generation of Backbone pSB6A1-AraC-lacZ
Miniprep of Amplified Parts
DH10ß containing I13453 and K20600 (for AraC) and backbone pSB1AK3 (lacZ) were grown ON at 37°C and minipreped.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| lacZ | 42.5 |
| AraC (5) | 26.5 |
| AraC (6) | 34.5 |
Restriction Digest
- AraC was cut with EcoRI-HF and SpeI buffered in NEB4
- LacZ was cut with XbaI and PstI buffered in NEB3
| Reagent | Amount [µl] |
|---|---|
| DNA | 22.5 |
| Enzymes | 1.5 each |
| Buffer (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 14.5 |
- Incubated for 1h at 37°C
- Purified with Qiagen Kit and diluted in 20 µl ddH2O
Result
| Sample | Concentration [ng/µl] |
|---|---|
| lacZ | 33.5 |
| AraC (5) | 19.5 |
| AraC (6) | 25.5 |
Ligation of pSB6A1, AraC and lacZ
| Reagent | Amount [µl] |
|---|---|
| pSB6A1 | 1.5 |
| lacZ | 4 |
| AraC | 3 |
| Ligase | 1.5 |
| Buffer | 2 |
| ddH2O | 8.5 |
- Incubated at RT for 45 min
- Chemical transformation of competent DH10ß
- Streaked on LB Amp plates
- Incubation ON at 37°C
Miniprep of pSB6A1-AraC-lacZ
- One colony each was picked and grown in 2 ml LB Amp ON
- Cultures were minipreped.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| pSB6A1-AraC-lacZ (5) | 57 |
| pSB6A1-AraC-lacZ (6) | 73 |
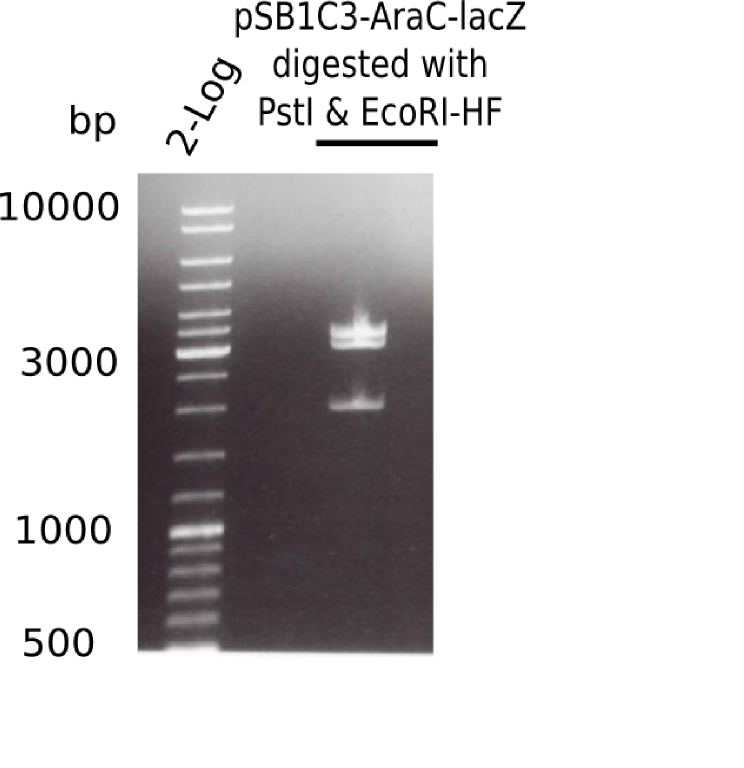
Restriction Digest of Backbone pSB6A1-AraC-lacZ
- AraC-lacZ was cut from pSB6A1-AraC-lacZ using PstI & EcoRI-HF
| Reagent | Amount [µl] |
|---|---|
| Miniprep DNA | 10 |
| Enzymes | 1.5 each |
| Buffer NEB 2(10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 27 |
- Incubated for 1 h at 37°C
- Purified with Qiagen kit and diluted in 20 µl ddH2O
Result
| Sample | Concentration [ng/µl] |
|---|---|
| pSB6A1-AraC-lacZ (5) D+P | 18 |
| pSB6A1-AraC-lacZ (6) D+P | 26 |
Ligation of AraC-lacZ with pSB1C3
| Reagent | Amount [µl] |
|---|---|
| DNA of pSB1C3 | 10 |
| DNA of AraC-lacZ (6) | 5 |
| Ligase | 1 |
| Buffer | 2 |
| ddH2O | 2 |
- Incubated at RT for 50 min
- Heat inactivation at 70°C for 5 min
- 10 µl were used for a chemical transformation in TOP10 and plated on LB Chlor
- Incubation ON at 37°C
Conlony-PCR Conditions BB.W3.A
| Reagent | psB1C3-AraC-lacZ (5) |
|---|---|
| Expected length [Kb] | ? |
| Template | Picked colony of psB1C3-AraC-lacZ (5) |
| Primer 10 µM fw | 0.5 µl |
| Primer 10 µM rev | 0.5 µl |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | - |
| ddH2O | 24 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 1:30 min |
| 1 | 4 | inf |
Result
Expected length:
There was no fragments visible.
- => Is picked coloniy negative or did entire ligation not work out?
Amplification of 1
20-05 - 26-05-13
DelH Fragment F1a
- The new primer arrived.
PCR Conditions F1a.W4.A
| Reagent | DelH F1a | DelH F1a |
|---|---|---|
| Expected length [Kb] | 5 | 5 |
| Template | 1 µl D. acidovorans | 1 µl D. acidovorans |
| Primer fw 10 µM | DN01 | DN01 |
| Primer rev 10 µM | DN08 | DN08 |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| DMSO | 2.5 µl | - |
| ddH2O | 20,5 | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 s |
| 62 | 5 s | |
| 72 | 2:00 min | |
| 1 | 72 | 2:30 min |
| 1 | 4 | inf |
Results
Expected band: 5 Kb
The gel shows a lot unspecific bands for both DelH F1a samples.
- => Alter PCR conditions.
DelH F1b
- The new primer arrived
PCR Conditions F1b.W4.A
| Reagent | DelH F1b |
|---|---|
| Expected length [Kb] | 5 |
| Template | 1 µl D. acidovorans |
| Primer fw 10 µM | DN07 |
| Primer rev 10 µM | DN02 |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | - |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 s |
| 62 | 5 s | |
| 72 | 2:00 min | |
| 1 | 72 | 2:30 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
The gel shows a lot unspecific bands for DelH F1b, but also the band at 5 Kb.
- => Band was cut and gel isolated.
Generation of Backbone pSB6A1-AraC-lacZ
Miniprep
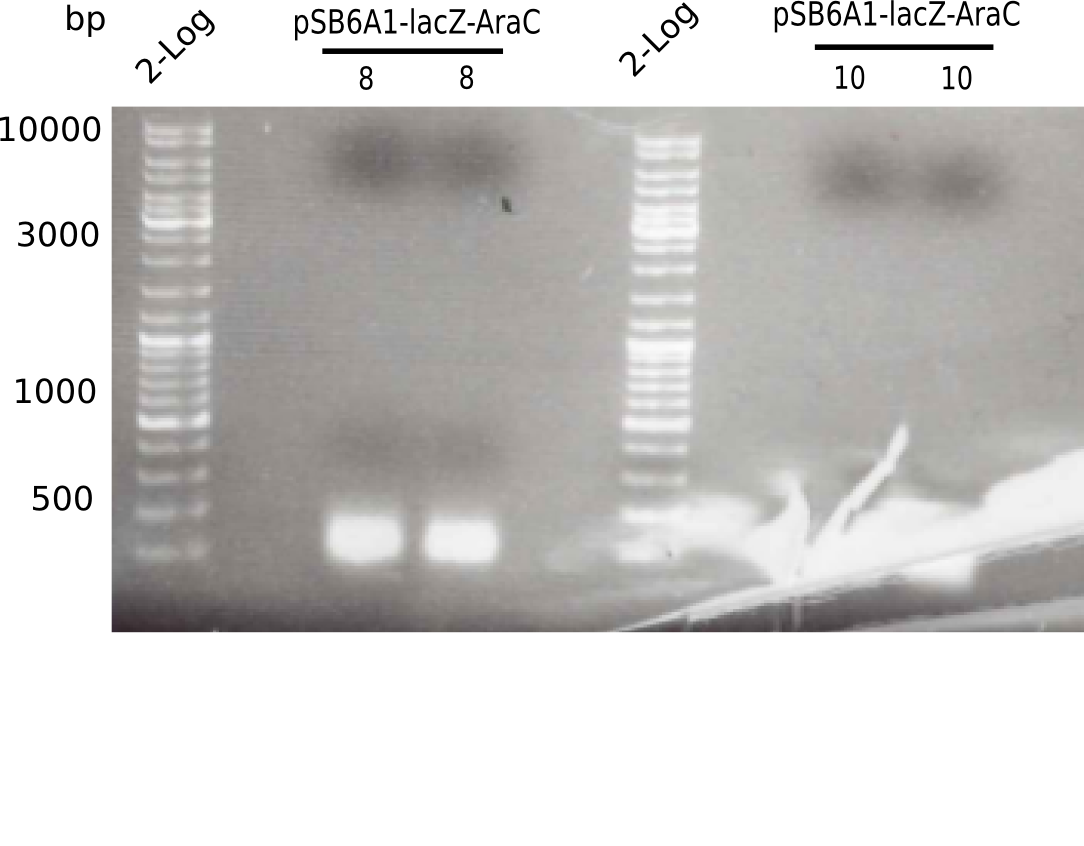
- From three different clones, 2ml of pSB6A1-AraC-lacZ (6) in LB Amp were minipreped and eluted in 30 µl ddH2O. Additionally, a glycerol stock was prepared.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| 8 - pSB6A1-AraC-lacZ (6) | 63 |
| 9 - pSB6A1-AraC-lacZ (6) | 61 |
| 10 - pSB6A1-AraC-lacZ (6) | 62 |
PCR Conditions BB.W4.A
| Reagent | 8 - pSB6A1-AraC-lacZ (6) | 9 - pSB6A1-AraC-lacZ (6) |
|---|---|---|
| Expected length [Kb] | 7.382 | 7.382 |
| Template | 1 µl of 1:10 diluted 8 | 1 µl of 1:10 diluted 9 |
| Primer 10 µM fw | 0.5 µl | |
| Primer 10 µM rev | 0.5 µl | |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| DMSO | - | - |
| ddH2O | 23 µl | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 s |
| 66↓ | 5 s | |
| 72 | 3:00 min | |
| 14 | 98 | 1 s |
| 63 | 5 s | |
| 72 | 3:00 min | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Results
Expected band: 7.4 Kb
The gel shows no bands.
- => Perform test restriction to test identity of plasmid.
Restriction Digest A
- Digest with PstI & XbaI of samples 8, 9 and 10 for checking identity of the construct
| Reagent | Amount [µl] |
|---|---|
| DNA | 10 |
| Enzymes | 1.5 each |
| Buffer NEB 3 (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 27 |
- Incubated for 1 h at 37°C
Result
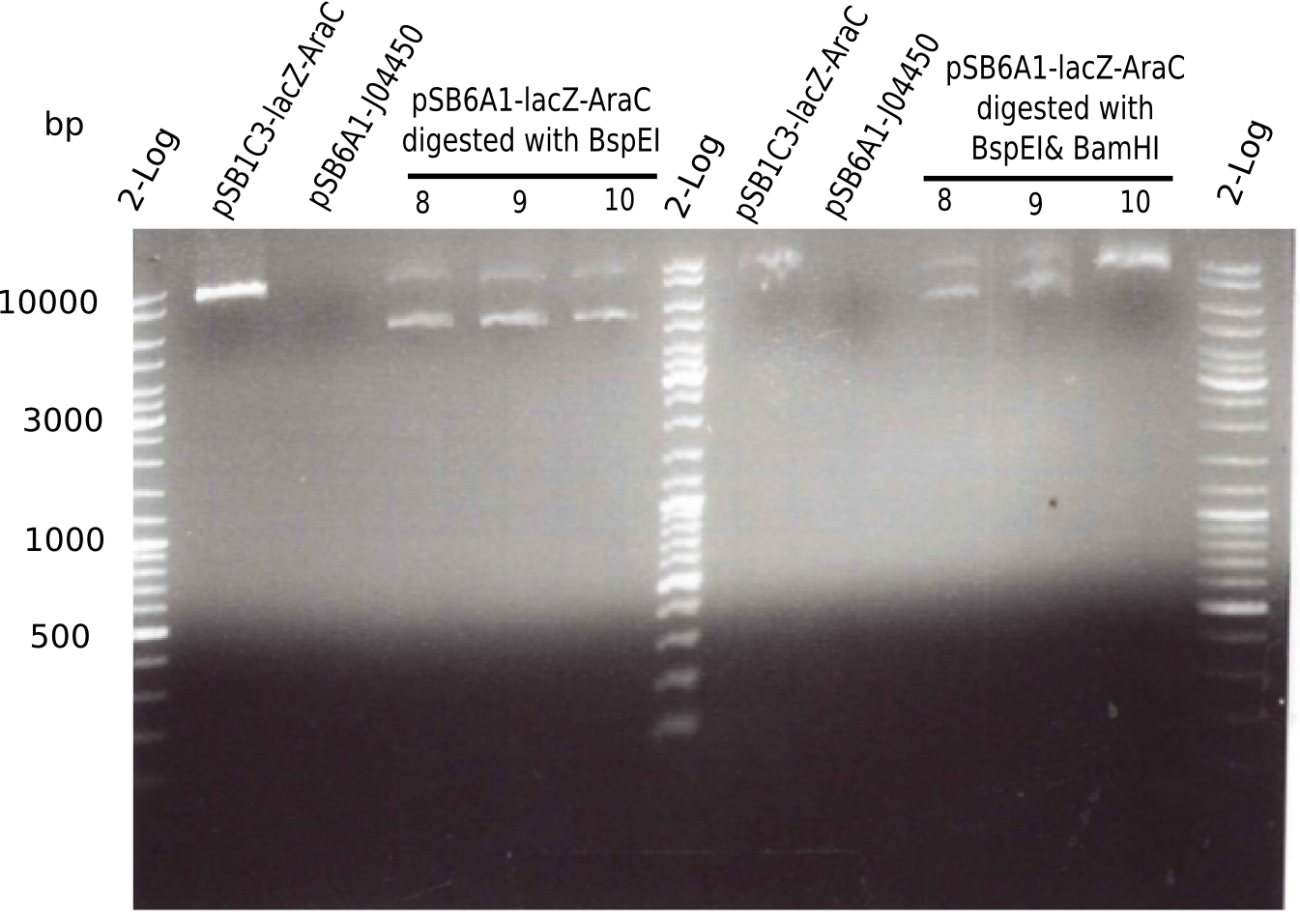
Expected bands: 3360 and 4022 bp
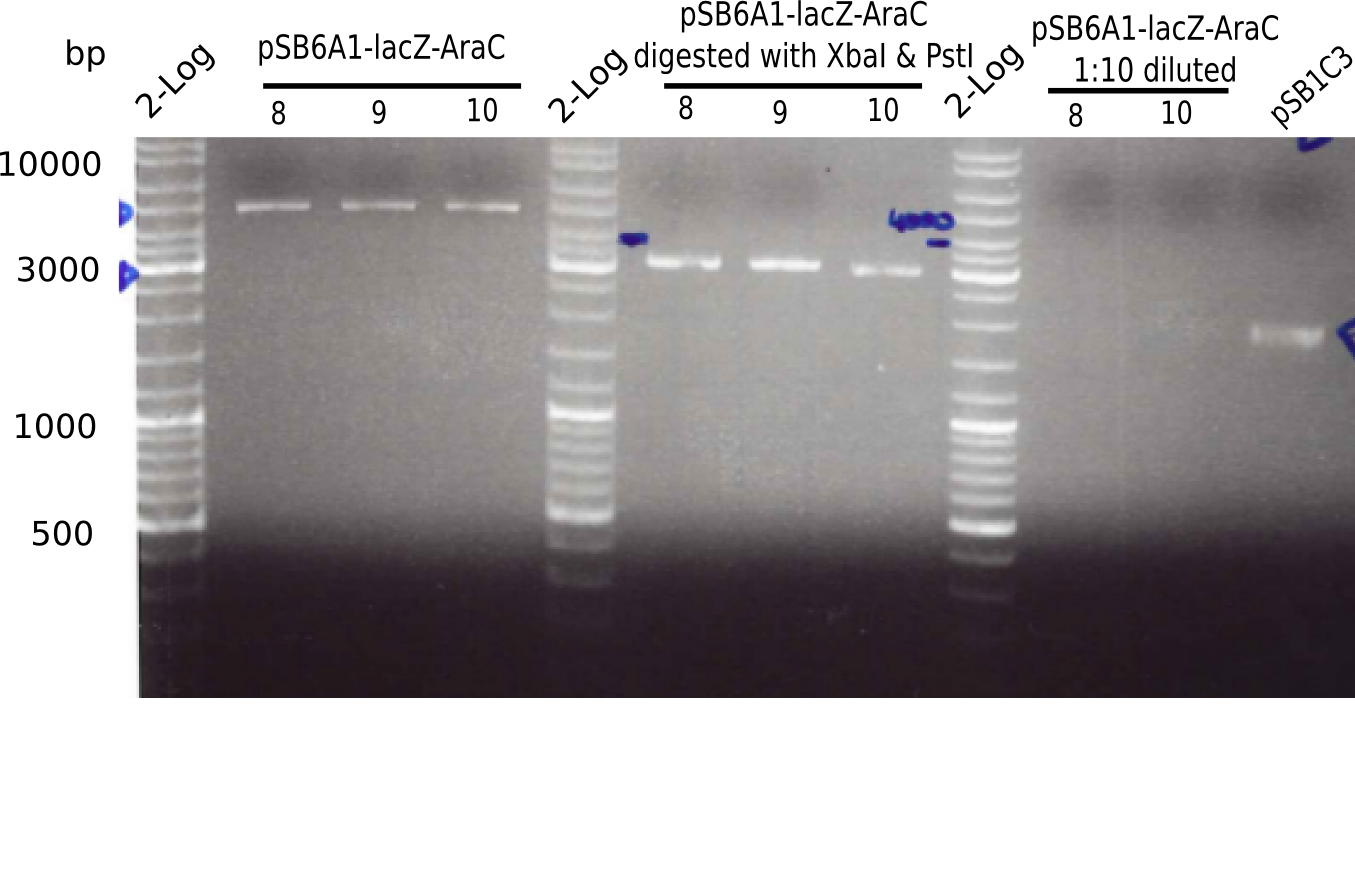
l1:2log ladder, l2: pSB6A1-AraC-lacZ (8), l3: pSB6A1-AraC-lacZ (9), l4: pSB6A1-AraC-lacZ (10),l5:2log ladder, l6: pSB6A1-AraC-lacZ (8) digested with XbaI & PstI, l7: pSB6A1-AraC-lacZ (9) digested with XbaI & PstI, l8: pSB6A1-AraC-lacZ (10)digested with XbaI & PstI, l9:2log ladder, l10: pSB6A1-AraC-lacZ (8) 1:10 diluted, l11: pSB6A1-AraC-lacZ (10) 1:10 diluted, l12: pSB1C3
The gel shows a band at 5 Kb.
- => Colonies 8, 9 and 10 are not the desired construct.
Restriction Digest B
- Digest with KpnI of samples 8, 9, 10 for checking length of the construct and KpnI and BamHI to check for identity.
| Reagent | Amount [µl] |
|---|---|
| pSB6A1-AraC-lacZ (8, 9, 10) | 5 |
| Enzymes | 1.5 KpnI |
| Buffer NEB 3 (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 33,5 |
| Expected bands | 7.381 bp |
| Reagent | Amount [µl] |
|---|---|
| pSB6A1-AraC-lacZ (8, 9, 10) | 5 |
| Enzymes | 1.5 KpnI & 1,5 BamHI |
| Buffer NEB 3(10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 32 |
| Expected bands | 4.607 bp & 2.781 bp |
Result
- Expected bands: 7.381 bp and 4.607 bp & 2.781 bp
- => Colonies 8, 9 and 10 are not desired construct. Pick new colony from pSB1C3-AraC(6)-lacZ and test digest.
Miniprep
A new colony was picked from pSB1C3-AraC(6)-lacZ plate and a mini prep was performed.
Result
pSB6A1-AraC(6)-lacZ construct : 94 [ng/µl]
Test Restriction Digest
The new miniprep as well as one of the wrong pSB6A1-AraC(6)-lacZ ones were restriction digested with EcoRI-HF and PstI.
| Component | Amount [µl] |
|---|---|
| DNA | 5 |
| BSA (10x) | 5 |
| NEB2 buffer (10x) | 5 |
| Enzymes (EcoRI-HF and PstI) | 1.5 each |
| ddH2O | 32 |
- Incubated 1 h at 37°C
Result
Expected bands: ? Kb
The gel showed again these three bands (2.070 and 3.360 and ~4 Kb)
- => Colonies 8, 9 and 10 are not the desired construct.
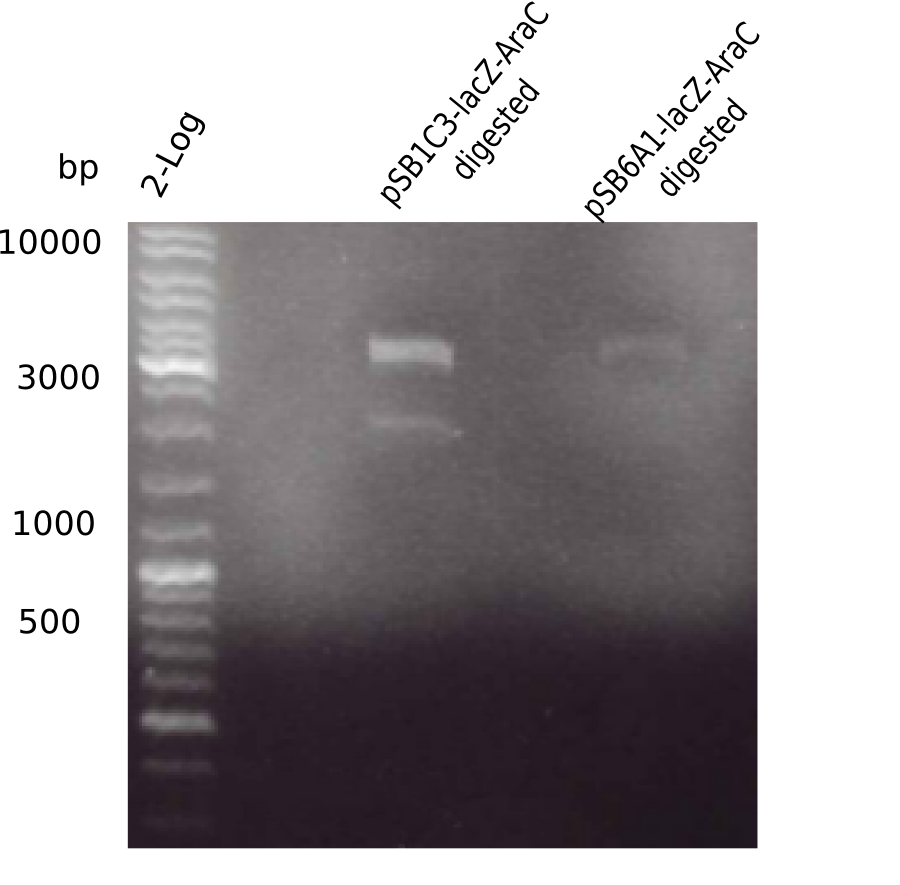
Generation of Backbone pSB1C3-AraC-lacZ
Test Restriction Digest
In order find out why pSB6A1-AraC-lacZ shows wrong restriction pattern, parental pSB1C3-AraC-lacZ was restriction digested using KpnI as well as KpnI & BamHI.
| Reagent | Amount [µl] |
|---|---|
| pSB1C3-AraC-lacZ | 5 |
| Enzymes | 1.5 KpnI |
| Buffer NEB 3(10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 33.5 |
| Expected band | 5,436 bp (linerized) |
| Reagent | Amount [µl] |
|---|---|
| pSB1C3-AraC-lacZ | 5 |
| Enzymes | 1.5 KpnI & 1.5 BamHI |
| Buffer NEB 3(10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 32 |
| Expected bands | 4.906 bp & 530 bp |
Result
Expected bands: 5.436 bp and 4.906 bp & 530 bp
- => pSB1C3-AraC(6)-lacZ is not desired construct. Pick new colony and test digest.
Miniprep
A new colony was picked from pSB1C3-AraC(6)-lacZ plate and a mini prep was performed.
Result
pSB1C3-AraC(6)-lacZ construct : 203 [ng/µl]
Test Restriction Digest
The new miniprep as well as one of the wrong pSB6A1-AraC(6)-lacZ ones were restriction digested with EcoRI-HF and PstI.
| Component | Amount [µl] |
|---|---|
| DNA | 3 |
| BSA (10x) | 5 |
| NEB2 buffer (10x) | 5 |
| Enzymes (EcoRI-HF and PstI) | 1.5 each |
| ddH2O | 34 |
- Incubated 1 h at 37°C
Result
Expected band: ? Kb
Gel showed again the three bands (2070 and 3360 and ~4 Kb).
- => pSB1C3-AraC(6)-lacZ is not desired construct. Start assembly all over again.
Test Restriction Digest pSB6A1-J04450
pSB6A1-J04450 was test digested to check for identity.
| Reagent | Amount [µl] |
|---|---|
| pSB6A1-J04450 | 10 |
| Enzymes | 1.5 KpnI |
| Buffer NEB 3 (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 28.5 |
| Expected band | 5,091 bp - Linearized |
| Reagent | Amount [µl] |
|---|---|
| pSB6A1-J04450 | 10 |
| Enzymes | 1.5 KpnI & 1.5 BamHI |
| Buffer NEB 3 (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 27 |
| Expected bands | 4,906 bp & 530 bp |
Result
| Sample | Expected bands with KpnI | Bands on gel with KpnI | Expected bands with KpnI & BamHI | Bands on gel with KpnI & BamHI |
|---|---|---|---|---|
| pSB1C3-AraC-lacZ | 5.436 Kb (linerized) | 10 Kb | 4.906 Kb & 530 Kb | no band |
| pSB6A1-J04450 | 5.091 Kb (linearized) | no band | 5.091 Kb (linearized) | no band |
| pSB6A1-AraC-lacZ (8, 9, 10) | 7.388 Kb (linearized) | 6 Kb & 10 Kb | 4.607 Kb & 2.781 Kb | 6Kb & 10 Kb |
This means:
- The chloramphenicol backbone has the right length.
- The digested chloramphenicol backbone is also ok.
- The digested fragment pSB6A1-AraC-lacZ (with PstI & EcoRI-HF) showed the 2 expected bands at 2,070 bp and 3,360 bp, but another band around ~4 Kb.
- => Something is not ok with this ligated construct and we have to start all over again.
Amplification of 1
27-05 - 02-06-13
Generation of pSB6A1
Miniprep
- Grow pSB6A1 from glycerol stock in 2 ml LB Amp ON at 37°C.
- Miniprep using miniprep kit from Qiagen and dilute in 30 µl ddH2O.
Result
Concentration 69.5 ng/µl
Digest
- The pSB6A1 was generated from the parts registry and cut with EcoRI & PstI
| Template DNA | BSA | NEBuffer2 | Enzymes | ddH2O | Total amount |
|---|---|---|---|---|---|
| 15 µl (69.5 ng/µl) | 5 µl | 5 µl | 2x 1.5 µl EcoRI-HF & PstI | 22 µl | 50 µl |
- The digest was incubated 45 min at 37°C.
Result
Expected band: 3985 bp (pSB6A1) and at 1106 bp (insert=J04450)
- => Expected bands are present. Backbone pSB6A1 was cut out and gel purified (c=4 ng/µl).
Generation of AraC Fragment
Gel Extraction
Gel extraction was performed with the gel extraction kit from Qiagen of gel slice (cut 24-05) and was eluted in 30 µl.
Result
The measured concentrations are shown in the table below and was stored at -20°C.
| Sample | Concentration [ng/µl] |
|---|---|
| AraC | 22.5 |
Generation of lacZ Fragment
Gel Extraction
Gel extraction was performed with the gel extraction kit from Qiagen of gel slice (cut 24-05) and was eluted in 30 µl.
Result
The measured concentrations are shown in the table below and was stored at -20°C.
| Sample | Concentration [ng/µl] |
|---|---|
| lacZ | 42.5 |
Generation of Backbone pSB6A1-AraC-lacZ
Ligation of pSB6A1 with AraC-lacZ
A ligation was performed of fragments for 1 h at RT.
| Sample | Concentration [ng/µl] | Amount for ligation [µl] |
|---|---|---|
| pSB6A1 | 4 | 12.5 |
| AraC-lacZ | 22.5 | 3.5 |
| T4-ligase | - | 1 |
| Buffer | - | 2 |
| ddH2O | - | 1 |
| Final volume | - | 20 |
- The ligase was inactivated by heat shock for 5 min at 70°C.
Transformation
- Afterwards, a transformation was performed with 15 µl of the ligation reaction in competent TOP10.
- Cells were streaked on LB Amp plates and incubated ON at 37°C.
Induction
- The following day, 5 white colonies were picked (because the red ones still have the mRFP insert J04450) and were incubated in 2 ml LB Amp with 20 µl Arabinose (10%) and 100 µl X-Gal (20 ng/µl).
Result
2 of the colonies turned blue
- => Perform miniprep.
Miniprep
Miniprep was performed according to Qiagen's instructions.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| pSB6A1-AraC-lacZ | 99 |
DelH F1a
PCR Conditions F1a.W5.A
| Reagent | DelH F1a | DelH F1a |
|---|---|---|
| Expected length [Kb] | 5 | 5 |
| Template | 1 µl D. acidovorans | 1 µl D. acidovorans |
| Primer fw 10 µM | DN01 | DN01 |
| Primer rev 10 µM | DN08 | DN08 |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| DMSO | 2.5 µl | - |
| ddH2O | 20,5 | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 |
| 62 | 5 | |
| 72 | 2:00 min | |
| 1 | 72 | 2:30 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
The gel shows a lot unspecific bands for both DelH F1a samples.
- => Fragment was cut and gel purified.
DelH F1b
PCR Conditions F1b.W5.A
| Reagent | DelH F1b |
|---|---|
| Expected length [Kb] | 5 |
| Template | 1 µl D. acidovorans |
| Primer fw 10 µM | DN07 |
| Primer rev 10 µM | DN02 |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | - |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:00 min | |
| 1 | 72 | 2:30 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
Gel shows one band at the expected length of 5 Kb, but also other side bands below and a huge, bright band at 2 Kb.
- => Band was cut and gel isolated.
DelH F2
PCR Conditions F2.W5.A
| Reagent | DelH F2 |
|---|---|
| Expected length [Kb] | 8 |
| Template | 1 µl glycerol stock |
| Primer 10 µM fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 0 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:45 min | |
| 1 | 72 | 2:45 min |
| 1 | 4 | inf |
Result
Expected length: 8 Kb
There was a specific band at 8 Kb.
- => Fragment was cut and gel extracted.
Amplification of 1
03-06 - 09-06-13
Generation of DelH plasmid pHM01
Elongation-PCR Conditions BB.W6.A
The restriction sites were added to pSB6A1-AraC-lacZ via PCR.
| Reagent | pSB6A1-AraC-lacZ |
|---|---|
| Expected length [Kb] | 7.4 |
| Template | 1 µl of 1:10 pSB6A1-AraC-lacZ miniprep |
| Primer fw 10 µM | DN05 |
| Primer rev 10 µM | DN06 |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | - |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 14 | 98 | 1 |
| 66↓ | 5 | |
| 72 | 2:00 min | |
| 16 | 98 | 1 |
| 66 | 5 s | |
| 72 | 2:00 min | |
| 1 | 72 | 2:30 min |
| 1 | 4 | inf |
Result
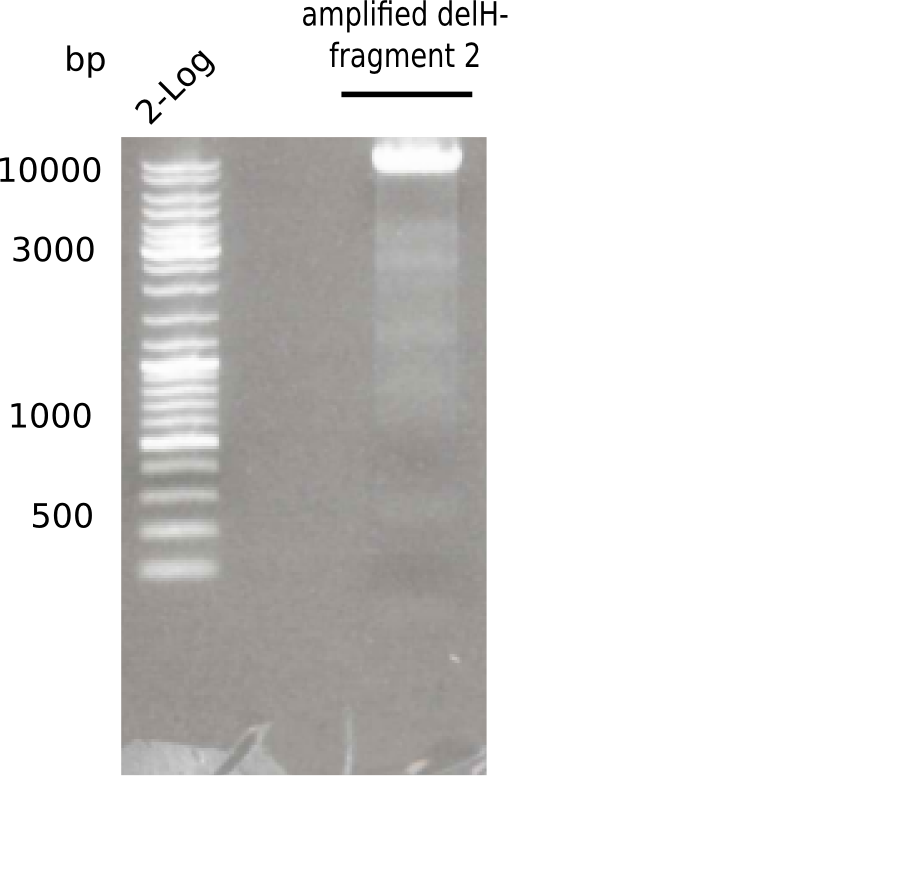
Expected band: 7.4 Kb
Gel shows expected band.
- => Band was cut and gel extracted.
Test Restriction Digest
Isolated fragment was test digested to confirm identity.
| Sample | Concentration [ng/µl] | Volume for digest [µl] | Enzyme [µl] | Buffer [µl] | ddH2O [µl] | Final volume [µl] |
|---|---|---|---|---|---|---|
| pSB6A1-AraC-lacZ | 99 | 5 | 1.5 EcoRI-HF & 1.5 PstI | 5 NEB2 & 5 BSA | 32 | 50 |
- The test digest was incubated 1 h at 37°C.
Result
Expected band: ~4 Kb & 3.4 Kb
Gel shows expected band.
- => Backbone is fine.
Restriction Digest of pSB6A1-AraC-lacZ for Ligation
| Sample | Concentration [ng/µl] | Volume for digest [µl] | Enzyme [µl] | Buffer [µl] | ddH2O [µl] | Final volume [µl] |
|---|---|---|---|---|---|---|
| pSB6A1-AraC-lacZ | 99 | 12.5 | 1.5 KpnI-HF & 1.5 PacI | 5 NEB4 & 5 BSA | 24.5 | 50 |
Restriction Digest was purified with Qiagen nucleotide removal kit.
Result
c=24 ng/µl
Restriction Digest of DelH F1a for Ligation
| Sample | Concentration [ng/µl] | Volume for digest [µl] | Enzyme [µl] | Buffer [µl] | ddH2O [µl] | Final volume [µl] |
|---|---|---|---|---|---|---|
| DelH F1a | ? | 25 | 1.5 EcoRI-HF & 1.5 PacI | 5 NEB4 & 5 BSA | 12 | 50 |
Restriction Digest was purified with Qiagen nucleotide removal kit.
Result
c=9 ng/µl
Restriction Digest of DelH F1b for Ligation
| Sample | Volume for digest [µl] | Enzyme [µl] | Buffer [µl] | ddH2O [µl] | Final volume [µl] |
|---|---|---|---|---|---|
| DelH F1b | 25 | 1.5 EcoRI-HF & 1.5 SalI-HF | 5 NEB4 & 5 BSA | 12 | 50 |
Restriction Digest was purified with Qiagen nucleotide removal kit.
Result
c=19 ng/µl
Restriction Digest of DelH F2 for Ligation
| Sample | Volume for digest [µl] | Enzyme [µl] | Buffer [µl] | ddH2O [µl] | Final volume [µl] |
|---|---|---|---|---|---|
| DelH F2 | 25 | 1.5 KpnI-HF & 1.5 SalI-HF | 5 NEB4 & 5 BSA | 12 | 50 |
Restriction Digest was purified with Qiagen nucleotide removal kit.
Result
c=9 ng/µl
Ligation of pHM01:DelH-pSB6A1-AraC-lacZ
Ligations was performed of all 4 fragments (digested and purified) for 1 h at RT in 2 different mixes.
| Sample | Concentration [ng/µl] | Volume for Ligation [µl] |
|---|---|---|
| pSB6A1-AraC-lacZ | 24 | 2 |
| DelH F1a | 9 | 6 |
| DelH F1b | 19 | 3 |
| DelH F2 | 9 | 6 |
| T4 ligase | - | 1 |
| Buffer | - | 2 |
| ddH2O | - | - |
| Final volume | - | 20 |
- The ligase was inactivated by heat shock for 5 min at 70°C.
- The reaction mix was purified with Qiagen nucleotide removal.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| 1 | 20 |
| 2 | 30 |
Electroporation
Two electroporations were performed.
- 20 ng
- 30 ng
- After an incubation time of 1 h in SOC-medium, cells were centrifuged and 10 µl plated on one LB Amp plate (with Arabinos and X-Gal) and 100 µl on another plate prepared with the same conditions.
- Plates were incubated ON at 37°C.
Result
White colonies grew on the plates.
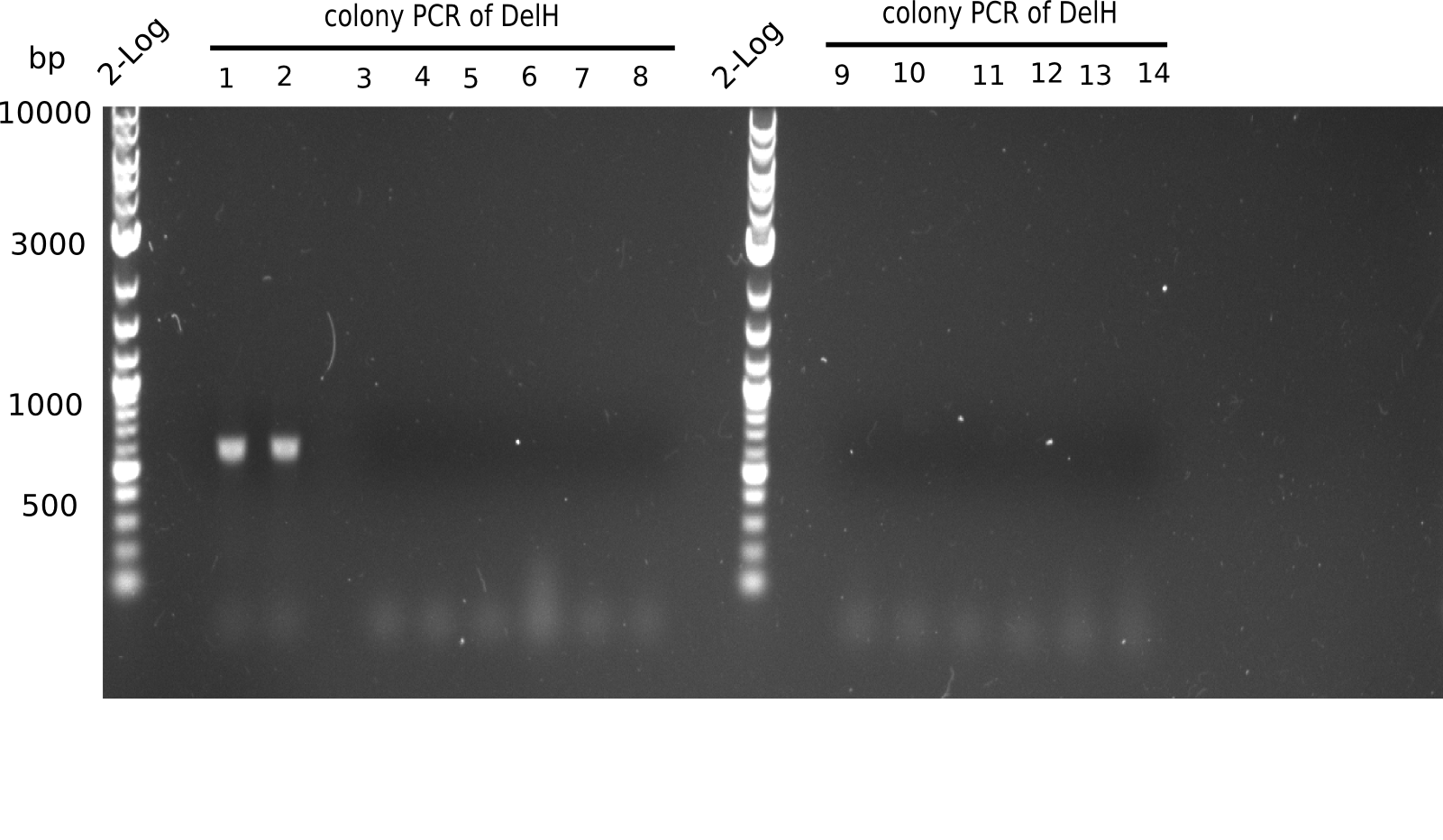
Colony PCR
6 colonies of plate E1 (colonies 1-6) and 6 colonies of plate E2 (7-12) as well as ligation mixes 1 & 2 were checked by PCR.
| Reagent | DelH colonies |
|---|---|
| Expected length [bp] | 663 |
| Primer fw 10 µM | 2 µl VF2 |
| Primer rev 10 µM | 2 µl DN07 |
| Taq Polymerase (2x) | 10 µl |
| ddH2O | 6 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 66 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 62 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected length: 663 bp.
None of the colonies shows a band. Ligation mixes 1 and 2 show the expected band at ~600 bp.
- => The ligation worked, but none of the correct plasmids were present in the analyzed colinies.
Amplification of 1
10-06 - 16-06-13
Characterization of DelH plasmid of 02-06
| Template | 15 x 1 µl of ligated plasmid |
|---|---|
| Expected length [bp] | 663 |
| Named | 1-15 |
| Primer fw 100 µM | 15 x 0.2 µl VF2 |
| Primer rev 100 µM | 15 x 0.2 µl DN07 |
| Dream-Taq Polymerase (2x) | 15 x 10 µl |
| ddH2O | 15 x 8.6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 94 | 180 |
| 12 | 94 | 20 |
| 66 (touchdown -0.5°C) | 20 | |
| 72 | 45 | |
| 18 | 94 | 20 |
| 62 | 20 | |
| 72 | 45 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Re-PCR of DelH fragment F1a & F1b
Result
Expected band: 5 Kb.
There are no specific bands at 5 Kb.
- => We therefore failed to reamplify DelH F1a and F1b from the PCR product.
- => Additionally, in the elctroporated colonies, there is no fragment F1a and F1b.
- => We need to design new forward primers for DelH F1a & F1b.
Amplification of DelH F1a
New Primers
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| DN09:DelH_f1_fw_long | 2013-06-11 | Amplification of DelH F1a from the genome: doesn't work | ATGGACCGTGGCCGCCTGCGCCAAATCG |
| DN10:DelH_f1_fw_short | 2013-06-11 | Amplification of DelH F1a from the genome: doesn't work | ATGGACCGTGGCCGCCTGC |
| DN11:DelH_f1_fw_short2 | 2013-06-11 | Amplification of DelH F1a from the genome: works!!!! | GCCGCCTGCGCCAAATCG |
| DN12:DelH_f1_PacI_fw_short | 2013-06-11 | Amplification of DelH F1a from the genome: doesn't work | ttttttaattaatcacacaggaaagtactagATGGACCGTGGCCGCCTGC |
PCR Conditions F1a.W7.A
| Reagent | DelH_f1_fw_long | DelH_f1_fw_short | DelH_f1_fw_short2 | DelH_f1_PacI_fw_short |
|---|---|---|---|---|
| Expected length [bp] | 5 Kb | 5 Kb | 5 Kb | 5 Kb |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock |
| Primer 10 µM fw | 2.5 µl DelH_f1_fw_long | 2.5 µl DelH_f1_fw_short | 2.5 µl DelH_f1_fw_short2 | 2.5 µl DelH_f1_PacI_fw_short |
| Primer 10 µM rev | 1 µl DelH_EcoRI_rev | 1 µl DelH_EcoRI_rev | 1 µl DelH_EcoRI_rev | 1 µl DelH_EcoRI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl | 25 µl | 25 µl |
| ddH2O | 19 µl | 19 µl | 19 µl | 19 µl |
Because the annealing temperature of DelH_f1_fw_long is 77,4°C and of the primer DelH_EcoRI_rev is 69,6°C, a 2-step PCR was performed.
| Cycles | Temperature DelH_f1_fw_long [°C] | Time [s] | Cycles | Temperature else [°C] | Time [s] | |
|---|---|---|---|---|---|---|
| 1 | 98 | 30 | 1 | 98 | 30 | |
| 30 | 98 | 5 | 30 | 98 | 5 | |
| - | - | 66 | 5 | |||
| 72 | 2:15 min | 72 | 2:15 min | |||
| 1 | 72 | 7 min | 1 | 72 | 7:00 min | |
| 1 | 4 | inf | 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb
There is a specific band at 5 Kb for DN11. The other primers show only unspecific products.
- =>The DN11 primer will be used. Using PCR conditions F1a.W7.A and short2 primer, the PCR was repeated to produce more product.
Amplification of 1
17-06 - 23-06-13
Amplification of DelH F1a
Elongation-PCR Conditions F1a.W8.A
| Reagent | Amount [µl] |
|---|---|
| Expected length [bp] | 5 |
| Template | 1 µl PCR-product (16-06) |
| Primer 10 µM fw | 2.5 µl DelH_f1_PacI_fw |
| Primer 10 µM rev | 2,5 µl DelH_EcoRI_rev |
| Phusion Master Mix (2x) | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 5 |
| 72 | 2:15 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb
Gels shows expected band.
- => Band was cut and gel extracted.
Restriction Digest
DelH F1a was restricted with EcoRI & PacI.
Afterwards it was purified with nucleotide removal kit and DNA concentration was measured.
Result
Expected band: 5 Kb
Gel shows expected band at ~5 Kb.
- => Restriction digest mix was purified by precipitation.
Purification of Restriction Digest
- Add 1 ml isopropanol
- Centrifuge 20 min full speed
- Discard supernatant
- Add 750 µl 70% ethanol
- Centrifuge 5 min full speed
- Discard supernatant
- Dry for 10 min
- Resuspend in 20 µl H2O
Result
DNA-concentration was measured at the Nanodrop c=21 ng/µl.
Re-PCR Conditions F1a.W8.A
In order to increase the product yield, F1a was amplified from the PCR fragment produced in week 7 using PCR conditions F1a.W7.A and short2 primer.
| Reagent | Amount [µl] |
|---|---|
| Expected length [bp] | 5 Kb |
| Template | 1 µl 1:10 dilution of purified DelH F1a (2.1 ng/µl) |
| Primer 10 µM fw | 2.5 µl DelH_f1_short2_fw |
| Primer 10 µM rev | 2.5 µl DelH_EcoRI_rev |
| Phusion Master Mix (2x) | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature A [°C] | Time [s] | Cycles | Temperature B [°C] | Time [s] | |
|---|---|---|---|---|---|---|
| 1 | 98 | 30 | 1 | 98 | 30 | |
| 30 | 98 | 5 | 30 | 98 | 5 | |
| - | - | 66 | 5 | |||
| 72 | 2:15 min | 72 | 2:15 min | |||
| 1 | 72 | 7 min | 1 | 72 | 7 min | |
| 1 | 4 | inf | 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb
Gel shows expected band at ~5 Kb.
- => Fragment was cut and gel isolated.
Generation of DelH plasmid 19-06
Ligation
Using 200 ng DNA
| Fragment | Size [Kb] | Concentration [ng/µl] | Volume [µl] for ligation |
|---|---|---|---|
| F1a | 5 | 261 (1:10 diluted => 26.1) | 1.4 |
| F1b | 5 | 19 | 1.6 |
| F2 | 8 | 9 | 4.5 |
| Backbone | 7.4 | 24 | 2 |
| Reagent | Volume [µl] for ligation |
|---|---|
| Ligase | 1 |
| Buffer | 2 |
| Total DNA | 10.3 |
| ddH2O | 7.3 |
Using 600 ng DNA
| Fragment | Size [Kb] | Concentration [ng/µl] | Volume [µl] for ligation |
|---|---|---|---|
| F1a | 5 | 261 (1:10 diluted => 26.1) | 8 |
| F1b | 5 | 19 | 7 |
| F2 | 8 | 9 | 2 |
| Backbone | 7.4 | 5 |
| Reagent | Volume [µl] for ligation |
|---|---|
| Ligase | 2 |
| Buffer | 4 |
| Total DNA | 22 |
| ddH2O | 22 |
Purification of Ligation
- Add 1 ml isopropanol
- Centrifuge 20 min full speed
- Discard supernatant
- Add 750 µl 70% ethanol
- Centrifuge 5 min full speed
- Discard supernatant
- Dry for 10 min
- Resuspend in 20 µl H2O
Electroporation
As described in methods Electroporation of E. coli DH10β
- One Eppi with DNA of ligation with 20 µl
- Second Eppi with DNA of ligation with 50 µl (more DNA)
- Streaked on LB Amp plates. Of each electroporated aliquot, one plate with 10 µl and another with 100 µl of the cells was spread.
- Incubation ON at 37°C.
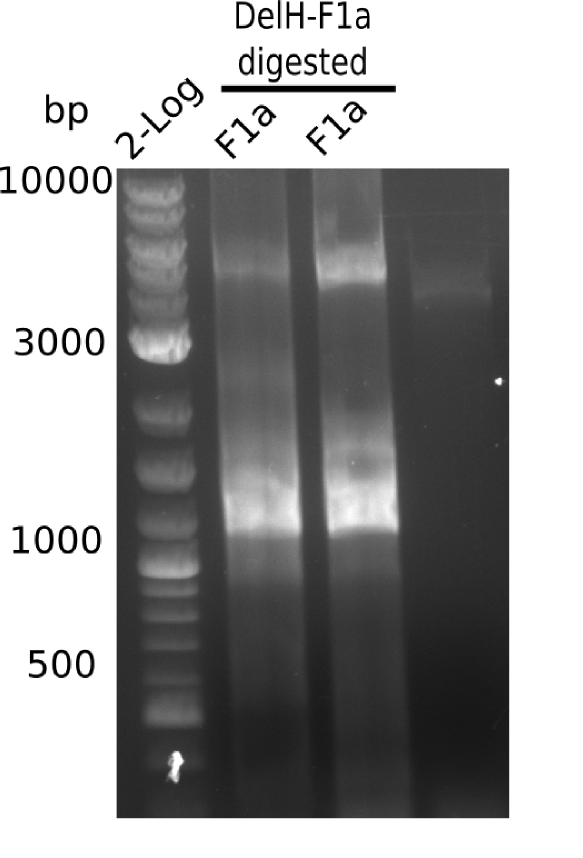
Characterization of DelH plasmid 19-06
Test Digest using SalI and PacI
| Reagent | Amount [µl] |
|---|---|
| DNA | 1 |
| SalI | 1 |
| PacI | 1 |
| NEB4-buffer | 2 |
| BSA | 2 |
| ddH2O | 13 |
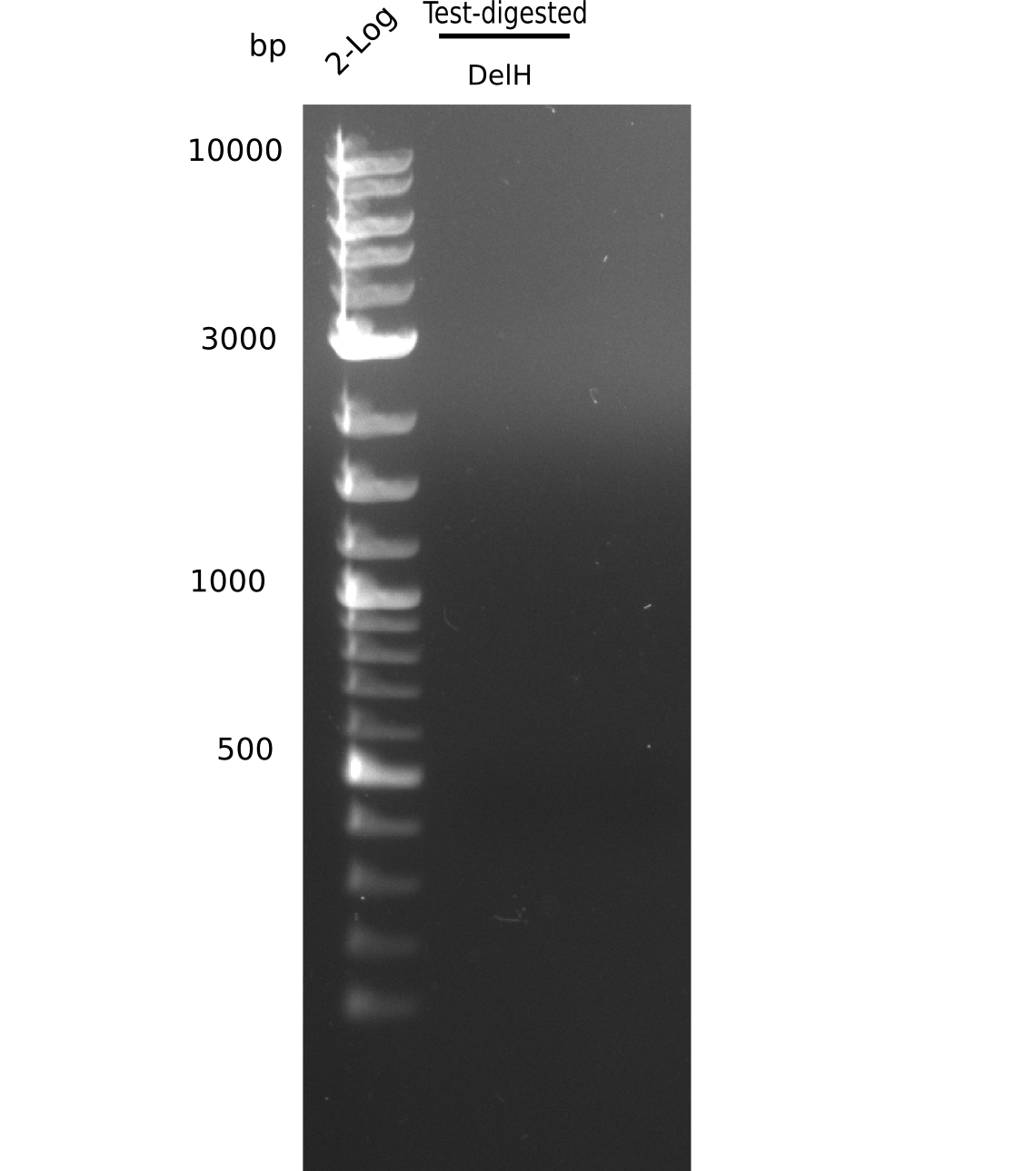
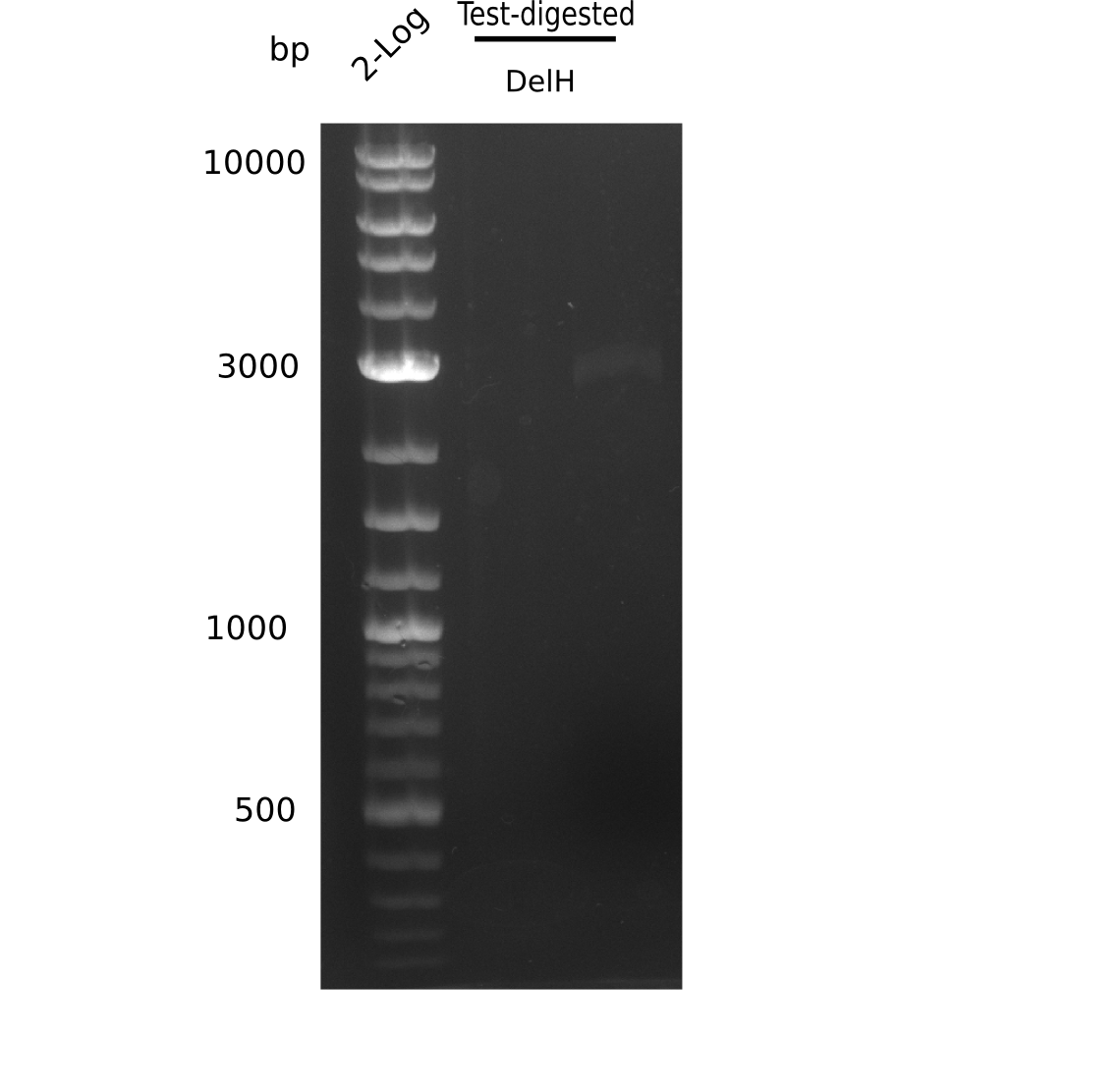
Result
Expected band: 10 Kb, 8 Kb, 7 Kb
There is nothing to see on gel, most probably due to too little amounts of DNA.
Colony-PCR
- Only few colonies (~10) grew on both 100 µl plates.
- Of 8 colonies from each 100 µl plate, a colony-PCR was performed.
- Inocculation of PCR colonies into 1 ml LB Amp-Ara-XGal ON at 37°C.
| Reagent | DelH colonies |
|---|---|
| Expected length [bp] | 663 |
| Primer fw 10 µM | 2 µl VF2 |
| Primer rev 10 µM | 2 µl DN07 |
| Dream-Taq Polymerase (2x) | 10 µl |
| ddH2O | 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Only colony 4 (second lane) shows expected band at ~600 bp. Yet, no blue color could be detected in ON culture, even in pellet (frozen for SDS Page).
- => This means, that our ligated plasmid was successfully transformed, but lacZ is not expressed.
Will run SDS-PAGE of transformed cultures and parental stock. Also inocculate cultures using higher concentrations of arabinose and X-Gal.
Stronger Induction of lacZ Expression
- Inocculated single colonies from day before into 5 ml LB Amp-Ara-XGal using 2x amounts of arabinose and X-Gal stock solutions (maybe concentrations of stocks are too low or promoters need stronger induction).
- Incubation ON at 37°C.
Result
No lacZ expression was observed.
Amplification of 1
24-06 - 30-06-13
Characterization of DelH plasmid 19-06
SDS Page
- Using NuPAGE Novex 10% Bis-Tris precast gel from Invitrogen with MOPS running buffer
- 3NuPAGE Novex 3-8% Tris-Acetate precast gel would have been more suitable considering expected 600 kDa DelH, but needed Tris-Acetate SDS buffer was not available.
- Resuspended pellets in 100 µl SDS buffer (from Linda)
- Boiled for 10 min @98°C
- Ran for 90 min, 180 V
- Stained for 40 min in Coomassie Buffer (50 ml acetic acid 100% + 225 ml ddH2O, 1.5 g Coomassie in 225 ml methanol, mix both)
- Washed in ddH2O multiple times
Result
None clear band at ~600 kDa visible. Maybe in highest amounts, but not reliable.
- => Probably no DelH expression in analyzed colonies.
Mini Prep
- Picked colonies from LB Amp plates into 5 ml LB Amp (colonies 4, 6 and 9)
- Incubation ON at 37°C
- 5 ml ON cultures were mini prepared and resuspended in 50 µl
Test Restriction Digest
- 5 µl of the mini prep were test digested
| Reagent | Volume [µl] |
|---|---|
| Miniprep DNA | 5 |
| CutSmart buffer | 2 |
| EcoRI-HF, PacI, KpnI-HF | 0.5 each |
| ddH2O | 10.75 |
- Incubation at 37°C for 1 h
Result
Expected band pattern: 5 kp, 7.3 kp, 13 kp
No clear bands visible.
- => DNA yield from mini prep was probably too low.
DNA Concentration
| Colony | DNA concentration [ng/µl] |
|---|---|
| 1 | 124.5 |
| 4 | 3.9 |
| 6 | 1.6 |
| 9 | 10.2 |
DNA measurement confirmed suspicion from test digest.
- =>Thus, mini prep and test digest have to be repeated
Repetion of Mini Prep
- Picked colonies from LB Amp plates into 5 ml LB Amp (colonies 4, 6 and 9) or from remaining ON culture (colony 1)
- Incubation ON at 37°C
- Colony 1 did not grow (also not on LB Amp plate), obviously lost plasmid
- Mini preped remaining 3, eluted in 35 µl ddH2O
DNA Concentration
| Colony | DNA concentration [ng/µl] |
|---|---|
| 4 | 38.5 |
| 6 | 42.0 |
| 9 | 49.0 |
Test Restriction Digest
- Digested entire mini prep (using remaining from first mini for colony 1) for 2 h at37°C
| Reagent | Volume [µl] |
|---|---|
| Mini prep | 42 |
| CutSmart | 5 |
| EcoRI-HF, PacI, KpnI-HF | 1 each |
Result
Expected bands: BB (7.3 Kb), DelH F2 (8 Kb), F1a + F1b (10 Kb)
Unexpected bands at 3 Kb, but not desired ones.
- => Colonies do not contain desired plasmid and entire ligation has to be repeated.
Amplification of DelH F1b
PCR Conditions F1b.W9.A
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl D. acidovorans stock 29-04-13 |
| Primer fw 10 µM | 0.5 µl DelH_EcoRI_fw 100 µM |
| Primer rev 10 µM | 0.5 µl DelH_f1_SalI_rev 100 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 30 s |
| 35 | 98 | 15 s |
| 68 | 5 s | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb, Loaded 1 µl of PCR
No correct band visible.
- => Why could we not reproduce the previous amplification?
PCR Conditions F1b.W9.B
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl D. acidovorans stock 29-04-13 |
| Primer fw 10 µM | 0.5 µl DelH_EcoRI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl DelH_f1_SalI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 30 s |
| 30 | 98 | 15 s |
| 68 | 15 s | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb, Loaded 1 µl of PCR
Again, expected band not there.
- => Adapt PCR conditions and ask for polymerase used last time, which is the one from the lab upstairs.
PCR Conditions F1b.W9.C
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl DelH F1b Fragment |
| Primer fw 10 µM | 0.5 µl DelH_EcoRI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl DelH_f1_SalI_rev 10 µM |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 10 s |
| 30 | 98 | 1 s |
| 68 | 5 s | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb, Loaded 1 µl of PCR
Highly specific band at 5 Kb, as well as additional smaller ones.
- => Band was cut. Run gel with remaining sample for gel extraction.
PCR Conditions F1b.W9.D
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl D. acidovorans stock 29-04-13 1:10 |
| Primer fw 10 µM | 0.5 µl DelH_EcoRI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl DelH_f1_SalI_rev 10 µM |
| Q5 Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 10 s |
| 30 | 98 | 1 s |
| 68 | 5 s | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: DelH F1b 5 Kb
Neither of the PCRs yield any product.
- => Therefore, Q5 is no option for us!
Amplification of DelH F2
PCR Conditions F2.W9.A
| Reagent | DelH 2 |
|---|---|
| Template | 1 µl D. acidovorans stock 29-04-13 |
| Primer fw 10 µM | 0.5 µl DelH_f2_SalI_fw 100 µM |
| Primer rev 10 µM | 0.5 µl DelH_f2_KpnI_rev 100 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 30 s |
| 35 | 98 | 15 s |
| 66 | 5 s | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 8 Kb
Unexpected bands at 3 Kb in lanes of DelH 2
- => Why could we not reproduce the previous amplification?
PCR Conditions F2.W9.B
| Reagent | DelH 2 |
|---|---|
| Template | 1 µl D. acidovorans stock 29-04-13 |
| Primer fw 10 µM | 0.5 µl DelH_f2_SalI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl DelH_f2_KpnI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 30 s |
| 12 | 98 | 15 s |
| 66 decrease by 0.5 | 15 s | |
| 72 | 3:30 min | |
| 18 | 98 | 15 s |
| 66 | 15 s | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 8 Kb
Unexpected bands at 1.5 Kb, 2.2 Kb, 4 Kb and 6 Kb. Desired 8 Kb band is present but hardly visible.
- => Adapt PCR conditions and ask for polymerase used last time, which is the one from the lab upstairs.
PCR Conditions F2.W9.C
| Reagent | DelH 2 |
|---|---|
| Template | 1 µl D. acidovorans stock 29-04-13 1:10 |
| Primer fw 10 µM | 0.5 µl DelH_f2_SalI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl DelH_f2_KpnI_rev 10 µM |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 10 s |
| 12 | 98 | 1 s |
| 66 decrease by 0.5 | 5 s | |
| 72 | 2:30 min | |
| 18 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 8 Kb
F2 shows specific band at 8 Kb and additional ones.
- => Band was cut. Run gel with remaining sample for gel extraction.
PCR Conditions F2.W9.D
| Reagent | DelH 2 |
|---|---|
| Template | 1 µl D. acidovorans stock 29-04-13 1:10 |
| Primer fw 10 µM | 0.5 µl DelH_f2_SalI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl DelH_f2_KpnI_rev 10 µM |
| Q5 Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 10 s |
| 12 | 98 | 1 s |
| 66 decrease by 0.5 | 5 s | |
| 72 | 2:30 min | |
| 18 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 8 Kb, Loaded 1 µl of PCR
Neither of the PCRs yield any product.
- => Therefore, Q5 is no option for us!
Amplification of Backbone pSB6A1-AraC-lacZ
PCR Conditions BB.W9.A
| Reagent | Backbone |
|---|---|
| Template | 1 µl Backbone (pSB6A1-AraC-lacZ c = 24 ng/µl) |
| Primer fw 10 µM | 0.5 µl AraCbb_KpnI_fw 100 µM |
| Primer rev 10 µM | 0.5 µl AraCbbPacI_rev2 100 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 30 s |
| 35 | 98 | 15 s |
| 66 | 5 s | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 7.3 Kb, Loaded 1 µl of PCR
No band visible, amplification failed.
- => Why could we not reproduce the previous amplification?
PCR Conditions BB.W9.B
| Reagent | BB |
|---|---|
| Template | 1 µl Backbone (pSB6A1-AraC-lacZ c = 24 ng/µl) |
| Primer fw 10 µM | 0.5 µl AraCbb_KpnI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl AraCbbPacI_rev2 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 30 s |
| 12 | 98 | 15 s |
| 66 decrease by 0.5 | 15 s | |
| 72 | 3:30 min | |
| 18 | 98 | 15 s |
| 66 | 15 s | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 7.3 Kb, Loaded 1 µl of PCR
Unexpected band at 2.2 Kb
- => Adapt PCR conditions and ask for polymerase used last time, which is the one from the lab upstairs.
PCR Conditions BB.W9.C
| Reagent | DelH 2 |
|---|---|
| Template | 1 µl pSB6A1-AraC-LacZ digested and purified 1:10 |
| Primer fw 10 µM | 0.5 µl AraCbb_KpnI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl AraCbbPacI_rev2 10 µM |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 10 s |
| 12 | 98 | 1 s |
| 66 decrease by 0.5 | 5 s | |
| 72 | 2:30 min | |
| 18 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 7.3 Kb, Loaded 1 µl of PCR
Gel does not show expected fragment.
- => Make sure right template was used. Is the restricted and purified fragment suitable?
PCR Conditions BB.W9.D
| Reagent | DelH 2 |
|---|---|
| Template | 1 µl pSB6A1-AraC-LacZ digested and purified 1:10 |
| Primer fw 10 µM | 0.5 µl AraCbb_KpnI_fw 10 µM |
| Primer rev 10 µM | 0.5 µl AraCbbPacI_rev2 10 µM |
| Q5 Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 10 s |
| 12 | 98 | 1 s |
| 66 decrease by 0.5 | 5 s | |
| 72 | 2:30 min | |
| 18 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 7.3 Kb, Loaded 1 µl of PCR
Neither of the PCRs yield any product.
- => Therefore, Q5 is no option for us!
Amplification of 1
01-07 - 07-07-13
Amplification of DelH F1b
PCR Conditions F1b.W10.A
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl DelH 1Fb Fragment |
| Primer fw 10 µM | 2.5 µl DelH_EcoRI_fw 10 µM |
| Primer rev 10 µM | 2.5 µl DelH_f1_SalI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 15 |
| 68 | 15 | |
| 72 | 5 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb
No correct bands visible.
- => Phusion is no option for us, not even for re-PCRs.
PCR Conditions F1b.W10.B
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl DelH_EcoRI_fw 10 µM |
| Primer rev 10 µM | 2.5 µl DelH_f1_SalI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 180 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb
- => F1b was not amplified.
Amplification of DelH F2
PCR Conditions F2.W10.A
| Reagent | DelH F2 |
|---|---|
| Template | 1 µl DelH 2 PCR product |
| Primer fw 10 µM | 2.5 µl DelH_f2_SalI_fw 10 µM |
| Primer rev 10 µM | 2.5 µl DelH_f2_KpnI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 15 |
| 66 decrease by 0.5 | 15 | |
| 72 | 8 min | |
| 12 | 98 | 15 |
| 66 | 15 s | |
| 72 | 8 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 8 Kb, Loaded 1 µl of PCR
Gel does not show correct band.
- => Phusion is no option for us, not even for re-PCRs.
PCR Conditions F2.W10.B
| Reagent | DelH F2 |
|---|---|
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl DelH_f2_SalI_fw 10 µM |
| Primer rev 10 µM | 2.5 µl DelH_f2_KpnI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 12 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 8 Kb, Loaded 50 µl of PCR
- => F2 was not amplified.
Amplification of Backbone
PCR Conditions BB.W10.A
| Reagent | Backbone |
|---|---|
| Template | 1 µl psB6A1+AraC+Lacz 1:10 |
| Primer fw 10 µM | 2.5 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 2.5 µl AraCbbPacI_rev2 |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 15 | |
| 72 | 8 min | |
| 12 | 98 | 15 |
| 66 | 15 | |
| 72 | 8 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 7.3 Kb, Loaded 1 µl of PCR
No product.
- => Phusion is no option for us, not even for re-PCRs.
PCR Conditions BB.W10.B
| Reagent | Backbone |
|---|---|
| Template | 1 µl psB6A1+AraC+Lacz 1:10 |
| Primer fw 10 µM | 2.5 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 2.5 µl AraCbbPacI_rev2 |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 12 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 7.3 Kb
- => BB was not amplified completely. It seems that there is no insert present or the conditions were not optimal. Repeat the PCR with other conditions.
PCR Conditions BB.W10.C
| Reagent | Backbone |
|---|---|
| Template | 1 µl psB6A1+AraC+Lacz 1:10 |
| Primer fw 10 µM | 2.5 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 2.5 µl AraCbbPacI_rev2 |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles-PCR | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:15 min | |
| 1 | 72 | 180 |
| 1 | 4 | inf |
Result
Expected bands: 7.3 Kb, Loaded 50 µl of PCR
Expected band is there, but also unspecific ones.
- => Correct band was cut and gel extracted.
Generation of DelH Plasmid
Current Status
| Fragment | Concentration [ng/µl] | Digested and Purified |
|---|---|---|
| F1a | 267 | check |
| F1b | 9.8 | - |
| F2 | 11 (gel extracted) and 369 (PCR purified) | - |
| pSB6A1-AraC-lacZ | 53.5 and 38.5 | - |
Restriction Digest of Fragments
| Fragment | Amount for Digest [ng] | Enzymes [µl] | Buffer [µl] |
|---|---|---|---|
| F1b | 441 |
EcoRI-HF: 1 | Cut smart 5.5 |
| F2 | 495 |
KpnI: 1 | Buffer 2.1: 5.5 |
| pSB6A1-AraC-lacZ | 1,599 |
KpnI: 1 | Buffer 1.1: 3.5 |
- 1 h at 37°C
Isopropanol Purification
| Fragment | Concentration after Digest & Purification [ng/µl] | Sequences |
|---|---|---|
| F1a | 267 |
File:Heidelberg PCR(DelH) restricted(KpnI+PacI) sequence.fasta.txt |
| F1b | 3.5 | (Entire) Sequence of DelH |
| F2 | 1.8 | |
| pSB6A1-AraC-lacZ | 7.3 |
File:Heidelberg PCR(pSB6A1-AraC-lacZ) digest.fasta.txt |
Confirmation of DNA Concentration
| Fragment | Concentration [ng/µl] | Date | µl on gel | Expected band size |
|---|---|---|---|---|
| F1a | 261 | 19-06-13 | 1 µl | 5 Kb |
| F1a | 261 | 19-06-13 | 2 µl | 5 Kb |
| F2 | 369 | 01-07-13 | 1 µl | 8 Kb |
Fragment F1a is not present, all others are fine.
- => Therefore, the next steps are:
- 1. Measurement of concentration = 40 ng/µl
- 2. PCR of the fragment F1a. As template, we use the purified PCR with the concentration of 40 ng/µl.
Restriction Digest of F2 and Backbone
| Fragment | Volume for digest [ng] | Enzymes [µl] | Buffer [µl] |
|---|---|---|---|
| 2 | 40*369 = 14.76 µg |
KpnI: 1 | Buffer 2.1: 4.8 |
| pSB6A1-AraC-lacZ | 40*38.5 = 1.54 µg |
KpnI: 1 | Buffer 1.1: 1.8 |
- 1h at 37°C
Purification of Restriction Digest
Using nucleotide removal kit (Qiagen), eluted with 30 µl ddH2O.
| Fragment | Concentration [ng/µl] | µl available |
|---|---|---|
| DelH F2 | 30 | 25 µl |
| pSB6A1-AraC-lacZ | -11 | 25 µl |
Amplification of DelH F1a
PCR Conditions F1a.W10.A
| Reagent | DelH F1a |
|---|---|
| Template | 0.5 µl of DelH F1a digested Fragment 1:5 (= 8 ng) |
| Primer fw 10 µM | 2.5 µl DelH_f1_PacI_fw |
| Primer rev 10 µM | 2.5 µl DelH_EcoRI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
Gel does not show any bands.
- => We conclude, that the fragments we expected to be F1a cannot be amplified by PCR. Thus, they were not amplified in the PCR used as template.
PCR Conditions F1a.W10.B
| Reagent | DelH F1a |
|---|---|
| Template | 1 µl ‘‘D. acidovorans’’ glycerol stock |
| Primer fw 10 µM | 2.5 µl DelH_f1_short2_fw |
| Primer rev 10 µM | 2.5 µl DelH_EcoRI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:00 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
See Confirmation of DNA Concentration
Amplification of DelH F1b
PCR Conditions F1b.W10.C
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl DelH F1b digested Fragment |
| Primer fw 10 µM | 2.5 µl DelH_EcoRI_fw |
| Primer rev 10 µM | 2.5 µl DelH_f1_SalI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
There is no band visible.
- => We conclude, that the fragments we expected to be F1b cannot be amplified by PCR. Thus, they were not amplified in the PCR used as template.
Generation of DelH Plasmid
Confirmation of DNA Concentration
| Fragment | Concentration (by Nanodrop) | µl on gel | µl loading dye | Digested and purified (V + A) | Date (of PCR or Purification) | Check on gel |
|---|---|---|---|---|---|---|
| F1a (PCR) | - | 1 | 9* 1x | - | 03-07-2013 (PCR) | + |
| F1a | 40 | 2 | 9* 1x | V + A | 19-06-2013 | - |
| F1b | 19 | 3 | 1* 6x | V + A | 19-06-2013 | - |
| F1b | 3.5 | 10 | 2* 6x | V + A | 02-07-2013 (V+A) | - |
| F2 | 0.5 | 10 | 2* 6x | V + A | - | |
| F2 | 30.5 | 2 | 10* 1x | V + A (Nucleotide Removal) | 03-07-2013 | + |
| F2 | (1:5 dilution of) 36.9 | 2 | 10* 1x | - | 02-07-2013 | + (BackUP) |
| pSB6A1-AraC-lacZ | 2.2 | 10 | 2* 6x | V + A | 02-07-2013 | + |
| pSB6A1-AraC-lacZ | (1:5 dilution of) 38.5 | 2 | 10* 1x | - | 02-07-2013 | + (BackUP) |
| pSB6A1-AraC-lacZ | -11 | 5 | 1* 6x | V + A (Nucleotide Removal) | 03-07-2013 | + |
DelH F1a (40 ng/µl) was the fragment we used for the first ligation. It is not visible, thus, there was not enough little DNA. Since the screening primer binds to this part of DelH, we could not identify any positive clone.
- => The negative samples (with too little DNA concentration) are discarded.
Amplification of DelH F1a
PCR Conditions F1a.W10.C
| Reagent | DelH F1a |
|---|---|
| Template | 1 µl DelH F1a Fragment 03-07-2013 |
| Primer fw 10 µM | 2.5 µl DelH_Pac_fw |
| Primer rev 10 µM | 2.5 µl DelH_EcoRI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles-PCR | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 120 | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Fragment was gel extracted.
Amplification of DelH F1b
PCR Conditions F1b.W10.D
| Reagent | DelH F1b | DelH F1b |
|---|---|---|
| Template | 1 µl D. acidovorans | 1 µl DelH F1b Fragment (19 ng/µl) |
| Primer fw 10 µM | 2.5 µl DelH_EcoRI_fw | 2.5 µl DelH_EcoRI_fw |
| Primer rev 10 µM | 2.5 µl DelH_f1_SalI_rev | 2.5 µl DelH_f1_SalI_rev |
| Phusion Ready Mix | 25 µl | 25 µl |
| ddH2O | 19 µl | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:15 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
No bands visible.
- => Fragment F1b could not be amplified from neither genomic DNA nor PCR fragment. Amplification will be repeated using DMSO in the mix.
PCR Conditions F1b.W10.E
| Reagent | DelH F1b | DelH F1b | DelH F1b | DelH F1b |
|---|---|---|---|---|
| Template | 1 µl D. acidovorans glycerol stock | 1 µl Fragment (19 ng/µl) | 1 µl D. acidovorans glycerol stock | 1 µl PCR Fragment (19 ng/µl) |
| Primer fw 10 µM | 1 µl DelH_EcoRI_fw | 1 µl DelH_EcoRI_fw | 1 µl DelH_EcoRI_fw | 1 µl DelH_EcoRI_fw |
| Primer rev 10 µM | 1 µl DelH_f1_SalI_rev | 1 µl DelH_f1_SalI_rev | 1 µl DelH_f1_SalI_rev | 1 µl DelH_f1_SalI_rev |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 7 µl | 7 µl | 6 µl | 6 µl |
| DMSO | - | - | 1 µl | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | |
| 72 | 2:15 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:15 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
- Using hot start 98°C
Result
Expected band: 5 Kb
Only PCR of fragment 19 ng/µl + DMSO was successful. Maybe there is another pale band at the second PCR of DelH F1b.
- => Repeat both PCRs.
Result
PCR using conditions F1b.W10.E was repeated.
Expected band: 5 Kb,Entire PCR mix was loaded.
PCR did not result in any fragments. Interestingly, both older PRCs resulted in a fragment at ~5 Kb and other lines, but the fragment from the genomic DNA is slightly larger.
- => Both fragments were cut and gel isolated, check correct length of F1b!
- => Perform PCR from the PCR fragment obtained from the genomic DNA.
Amplification of DelH F2
PCR Conditions F2.W10.C
- 2x 50 µl
| Reagent | DelH F2 |
|---|---|
| Template | 1 µl Fragment "BackUP" |
| Primer fw 10 µM | 2.5 µl DelH_f2_SalI_fw |
| Primer rev 10 µM | 2.5 µl DelH_f2_KpnI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 8 Kb
Fragment 2 was successfully amplified.
- => It was cut and gel purified.
Preparation of new Template DNA
- D. acidovorans glycerol stock was streaked onto LB plates and incubated ON at 37°C.
- D. acidovorans did not grow, since LB media is not appropriate for them. Prepare new ACM media and plates and streak D. acidovorans.
Amplification of Backbone
PCR Conditions BB.W10.D
- 2x 50 µl
| Reagent | Backbone |
|---|---|
| Template | 1 µl Fragment "BackUP" |
| Primer fw 10 µM | 2.5 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 2.5 µl AraCbbPacI_rev2 |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 7.3 Kb
No band visible.
- => Repeat using DMSO.
PCR Conditions BB.W10.E
| Reagent | Backbone | Backbone | Backbone |
|---|---|---|---|
| Template | 1 µl of BackUP | 1 µl V+A 2,2 ng/µl template | 1 µl V+A 2,2 ng/µl template |
| Primer fw 10 µM | 1 µl AraCbb_KpnI_fw | 1 µl AraCbb_KpnI_fw | 1 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 1 µl AraCbbPacI_rev2 | 1 µl AraCbbPacI_rev2 | 1 µl AraCbbPacI_rev2 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl |
| ddH2O | 7 µl | 7 µl | 6 µl |
| DMSO | - | - | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
- Using hot start (in new cycler)
Result
Expected band: 7.3 Kb
Gel does not show the expected fragment.
- => Next PCR for the Backbone should be done with longer elongation time and variations: think about using the a new colony of plate.
PCR Conditions BB.W10.F
| Reagent | Backbone | Backbone | Backbone | Backbone |
|---|---|---|---|---|
| Template | 1 µl Fragment 24 ng/µl | 1 µl of Fragment pSB6A1 gel-purified 3-6 89 ng/µl | 1 µl of digested Fragment 3-7 -11 ng/µl | 1 µl of purified Fragment Isoprop 2.2 ng/µl |
| Primer fw 10 µM | 1 µl DelH_f2_SalI_fw | 1 µl AraCbb_KpnI_fw | 1 µl DelH_f2_SalI_fw | 1 µl AraCbb_KpnI_fw 1 |
| Primer rev 10 µM | 1 µl DelH_f2_KpnI_rev | 1 µl AraCbbPacI_rev2 | 1 µl DelH_f2_KpnI_rev | 1 µl AraCbbPacI_rev2 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 7 µl | 7 µl | 7 µl | 7 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 180 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 180 | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 7.3 Kb,Entire PCR mix loaded (pSB6A1-AraC-lacZ)
Only PCRs from Fragment 24 ng/µl and Fragment 3-6 produced a nice fragment.
- => Bands were cut and gel isolated.
- => PCR amplification from restriction digested fragment did not work! This is unexpected, because actually, it should. Maybe exclude the previous restriction digested fragments "V+A" from ligation.
Preparation of new Template DNA
- psB6A1-LacZ-AraC: 2 ml LB Amp were inocculated from the plate and incubated ON at 28°C (all other shakers occupied).
- Mini prep was performed.
- 1 µl mini prep DNA was electroporated into E.coli DH10ß according to Electroporation of E. coli DH10β
Amplification of 1
08-07 - 14-07-13
Amplification of DelH F1b
PCR Conditions F1b.W11.A
| Reagent | DelH F1b - from purified F1b 05-07 (genome amplified) | DelH F1b - from purified F1b 05-07 (genome amplified) |
|---|---|---|
| Template | 1 µl of PCR 1b (19 ng/µl) | 1 µl D. acidovorans |
| Primer fw 10 µM | 1 µl DelH_EcoRI_fw | 1 µl DelH_EcoRI_fw |
| Primer rev 10 µM | 1 µl DelH_f1_SalI_rev 10 | 1 µl DelH_f1_SalI_rev |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 7 µl | 6 µl |
| DMSO | - | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:15 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
There is a specific band, but not at 5 Kb.
- => We cannot amplify F1b from a PCR fragment. Use fresh bacteria instead.
Preparation of fresh ACM media
- Mix ingrediants according to Acidovorax complex medium
- Autoclave
- pH adjust to 7,3
- Mix 500 ml media and 6 g agar
- Autoclave again
- Pour plates
Preparation of fresh D. acidovorans
- Inocculate 5 ml ACM media with D. acidovorans
- Incubate ON
- Prepare glycerol stock
- Streak some of culture on plates
PCR Conditions F1b.W11.B
| Reagent | DelH F1b | DelH F1b |
|---|---|---|
| Template | Fresh colony of plate by DN from 10-07 | Fresh colony of plate by DN from 10-07 |
| Primer fw 10 µM | 1 µl delH_EcoRI_fw | 1 µl delH_EcoRI_fw |
| Primer rev 10 µM | 1 µl delH_f1_SalI_rev | 1 µl delH_f1_SalI_rev |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 7 µl | 6 µl |
| DMSO | - | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 5.4 Kb
There is a weak band at the probe 1b WITH DMSO above 5 Kb.
- => Start a new PCR with 50 µl with the same conditions.
PCR Conditions F1b.W11.C
| Reagent | DelH 1b |
|---|---|
| Template | Fresh colony of plate by DN from 10-07 |
| Primer fw 10 µM | 2.5 µl delH_EcoRI_fw |
| Primer rev 10 µM | 2.5 µl delH_f1_SalI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 17.5 µl |
| DMSO | 2.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb.
The expected band at 5 Bk is weak, but visible. Note that there is sample left in the pocket.
- => The fragment was cut and gel extracted.
Generation of DelH plasmid
Restriction Digest
Of fragments F1a, F1b, F2 and backbone.
| Fragment | DNA [µl] | ddH2O [µl] | Enzymes [µl] | BSA [µl] | Buffer 4 [µl] |
|---|---|---|---|---|---|
| F1a (1) | 30 | 7 | 1.5 Pac & EcoRI-HF each | 5 | 5 |
| F1a (2) | 30 | 7 | 1.5 Pac & EcoRI-HF each | 5 | 5 |
| F1b (11.7 - 20 µl) | 20 | 17 | 1.5 SalI-HF & EcoRI-HF each | 5 | 5 |
| F1b (11.7 - 50 µl) | 20 | 17 | 1.5 SalI-HF & EcoRI-HF each | 5 | 5 |
| F2 | 20 | 17 | 1.5 SalI-HF & KpnI-HF each | 5 | 5 |
| BB (6-7 of IK - 14 ng/µl) | 20 | 17 | 1.5PacI & KpnI-HF each | 5 | 5 |
| BB (6-7) | 20 | 17 | 1.5 PacI & KpnI-HF each | 5 | 5 |
Result
Loaded 5 µl of restriction digested fragments on gel. The result stands in the table below:
| Fragment | Band | Conclusion | Next step |
|---|---|---|---|
| F1a purified on 12-07 | strong bands at 1, 2 and 3 Kb weak band at wanted 5 KB | PCR purification of F1a step was not the right purifying step | load complete probe on gel and gel extract |
| F1b purified on 12-07 | weak band at 5 Kb | maybe only low concentration | run PCR again |
| F2 | no band | maybe too low concentration | run PCR again |
| BB | weak band at 7.4 Kb | maybe only low concentration | run PCR again |
Gel Extraction of DelH F1a
Expected band: 5 KB, loaded entire F1a purified on 12-07 on gel
No band visible.
- => Gel extract earlier PCR from 06-07.
Expected band: 5 Kb
PCR shows expected band.
- => F1a was cut and gel extracted.
Amplification of DelH F1b
PCR Conditions F1b.W11.D
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl F1b digested 12-07 |
| Primer fw 10 µM | 1 µl delH_EcoRI_fw |
| Primer rev 10 µM | 1 µl delH_f1_SalI_rev |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 6µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0,5°C) | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 5.4 Kb
PCR did not yield the specific band.
- => Use altered PCR conditions.
Amplification of DelH F2
PCR Conditions F2.W11.A
| Reagent | DelH F2 |
|---|---|
| Template | 1 µl F2 digested 12-07 |
| Primer fw 10 µM | 1 µl delH_f2_SalI_fw |
| Primer rev 10 µM | 1 µl delH_f2_KpnI_rev |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 5 s |
| 30 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 8 Kb
PCR does not show the specific band.
- => Further optimization.
Amplification of Backbone
PCR Conditions BB.W11.A
| Reagent | Backbone |
|---|---|
| Template | 1 µl backbone digested 12-07 |
| Primer fw 10 µM | 1 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 1 µl AraCbb_PacI_rev2 10 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 8 Kb
PCR does not show the specific band.
- => Repeat using fresh backbone DNA from miniprep.
Miniprep
- Inocculation of 10 ml LB Amp with DH10ß (pSB6A1-AraC-lacZ)
- Perform mini prep
PCR Conditions BB.W11.B
| Reagent | Backbone |
|---|---|
| Template | 1 µl Miniprep A of 10-07 |
| Primer fw 10 µM | 1 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 1 µl AraCbb_PacI_rev2 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 7 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 (touchdown -0.5°C) | 5 | |
| 72 | 3:00 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results
Expected band: 7.4 Kb, loaded miniprep and PCR product.
Miniprep shows weak band at ~7.4 Kb. PCR does not show expected band.
- => Prepare new mediprep of backbone (pSB6A1-AraC-lacZ)
Gibson Assembly Strategy
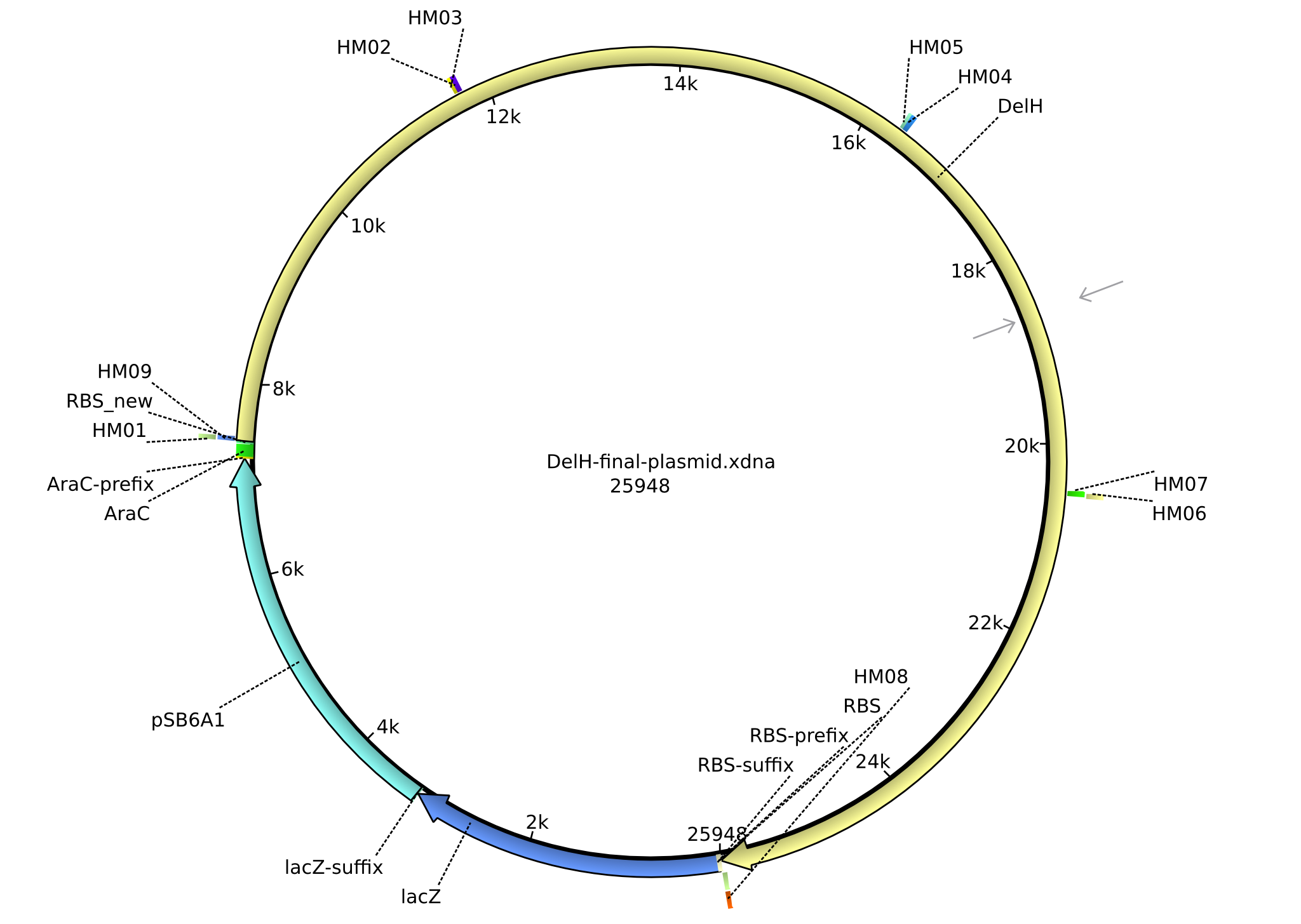
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| DN01:DelH_f1_PacI_fw | 03-05-2013 | Amplification of DelH F1, with RBS and adding PacI restriction site | TTTT TTAATTAA TCACACAGGAAAGTACTAG ATGGACCGTGGCCGCCTGC GCCAAATCG |
| DN02:DelH_f1_SalI_rev | 03-05-2013 | Amplification of DelH F1 until SalI restriction site | TTTT GTCGACCAACACCTGTGCCTGC |
| DN03:DelH_f2_SalI_fw | 03-05-2013 | Amplification of DelH F2 starting at SalI restriction site | TTTT GTCGACTGGATGGAGCCTGGTGAAAG |
| DN04:DelH_f2_KpnI_rev | 03-05-2013 | Amplification of DelH F2, adding KpnII restriction site | TTTT GGTACC TCAGTCCAGCGCGTACTCCAG |
| DN05:AraCbb_KpnI_fw | 03-05-2013 | Amplification of backbone for DelH (pSB6A1-AraC-lacZ), adding KpnI site | TTTT GGTACC AAAAGAGGAGAAATACTAGATGACCATG |
| DN08:AraCbb_PacI_rev | 03-05-2013 | Amplification of backbone for DelH (pSB6A1-AraC-lacZ), adding PacI site | TTTT TTAATTAA GCTAGCCCAAAAAAACGGGTATG |
Amplification of 1
15-07 - 21-07-13
Preparation of Primers
- All primers were 1:10 diluted
- Overview on annealing temperatures
| Primer | short2 | HM02 | HM03 | HM04 | HM05 | HM06 | HM07 | HM08 | HM09 | HM10 |
| Annealing temperature [°C] | 63 | 72.6 | 69.1 | 55.2 | 68.2 | 66.4 | 69.6 | 65.2 | 51.6 / 73.6 | 60.9 / 71.3 |
Preparation of fresh D. acidovorans
- Liquid culture: 2x 5 ml ACM medium was inocculated with picked colonies of D. acidovorans from plate (made by DN)
- Glycerol stock: was prepared from liquid culture
Amplification of DelH G1
PCR Conditions G1.W12.A
| Reagent | DelH G1 | DelH G1 |
|---|---|---|
| Template | Fresh colony of plate (by DN) | Fresh colony of plate (by DN) |
| Primer fw 10 µM | short2 | short2 |
| Primer rev 10 µM | HM04 | HM04 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 7 µl | 6 µl |
| DMSO | - | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 63 (touchdown -0.5°C) | 5 | |
| 72 | 3:30 min | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 8.961 Kb
No bands visible.
- => Fragment 1 was not amplified. Next step is amplifying fragment 1 in two parts G1a & G1b.
Amplification of DelH G1a
PCR Conditions G1a.W12.A
| Reagent | DelH G1a | DelH G1a |
|---|---|---|
| Template | Fresh colony of plate (by DN) | Fresh colony of plate (by DN) |
| Primer fw 10 µM | short2 | short2 |
| Primer rev 10 µM | HM02 | HM02 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 7 µl | 6 µl |
| DMSO | - | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 63 (touchdown -0.5°C) | 5 | |
| 72 | 3:30 min | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected Band: 4.309 Kb
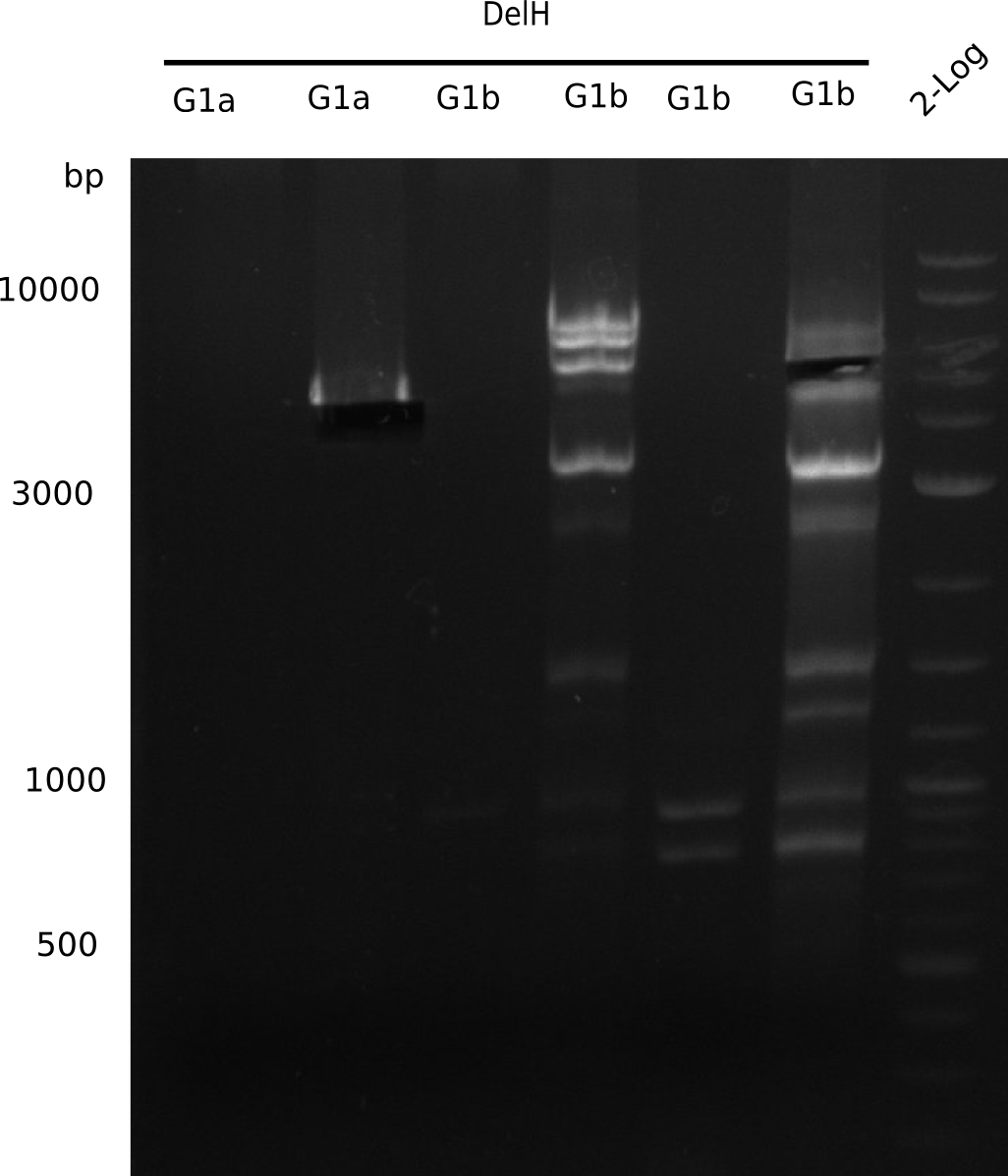
l1:F1a without DMSO,l2:F1a with DMSO, l3:F16 without DMSO at an anealing temperature 64°C,l5:F16 with DMSO at an anealing temperature 64°C,l6:1b-57 (without DMSO), l7: 1b-57 (with DMSO),l9 2 loga ladder,l10 Mediprep A of BB,l11 Mediprep B of BBF1a with DMSO
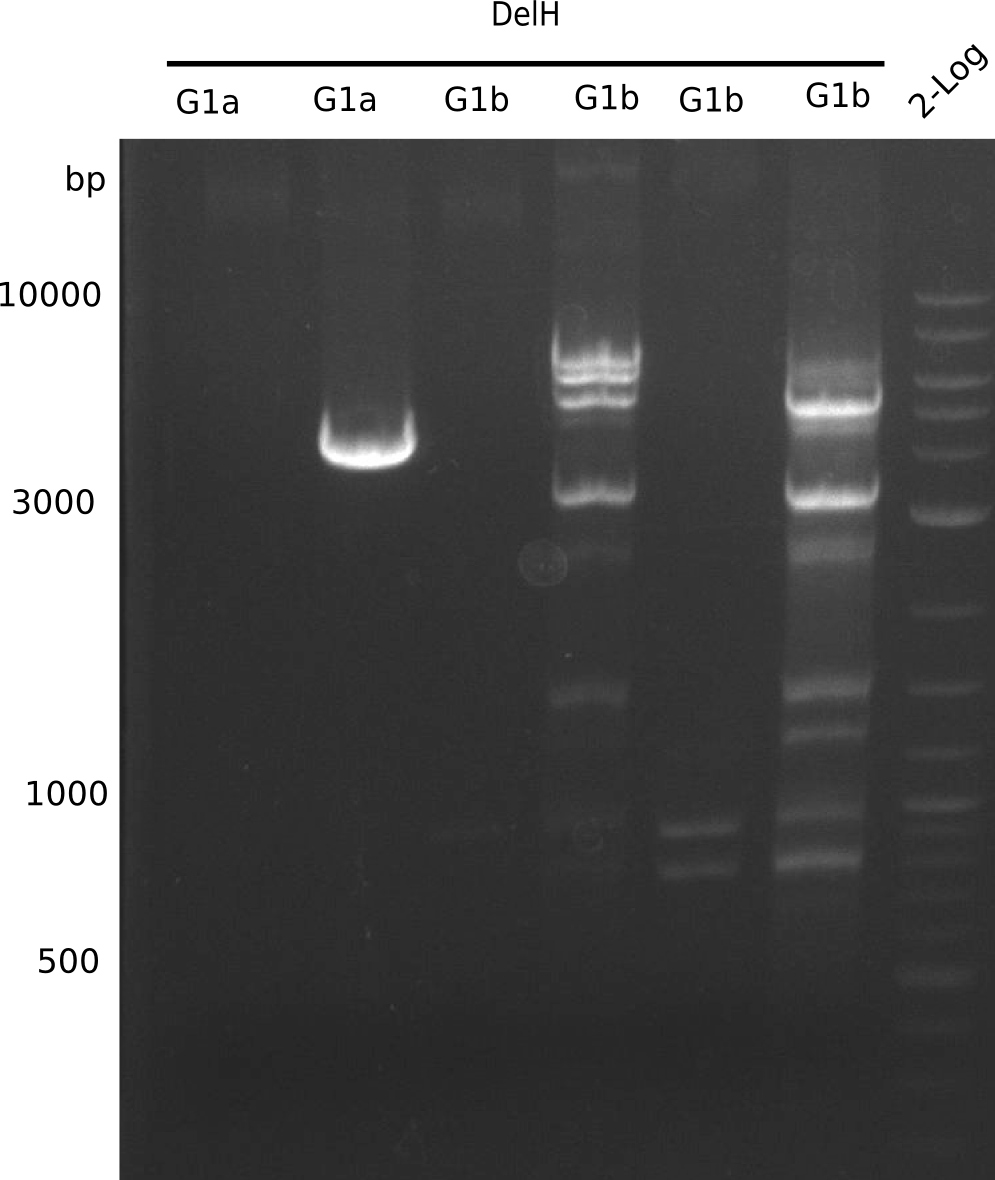
l1:F1a without DMSO,l2:F1a with DMSO, l3:F16 without DMSO at an anealing temperature 64°C,l5:F16 with DMSO at an anealing temperature 64°C,l6:1b-57 (without DMSO), l7: 1b-57 (with DMSO),l9 2 loga ladder,l10 Mediprep A of BB,l11 Mediprep B of BBF1a with DMSO
PCR using DMSO looks well.
- => Fragment was cut and gel extracted.
Amplification of DelH G1b
PCR Conditions G1b.W12.A and B
| Reagent | DelH G1b | DelH G1b |
|---|---|---|
| Template | Fresh colony of plate (by DN) | Fresh colony of plate (by DN) |
| Primer fw 10 µM | HM03 | HM03 |
| Primer rev 10 µM | HM04 | HM04 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 7 µl | 6 µl |
| DMSO | - | 1 µl |
| Cycles | Temperature A [°C] | Time [s] | Cycles | Temperature B [°C] | Time [s] | |
|---|---|---|---|---|---|---|
| 1 | 98 | 5 | 1 | 98 | 5 | |
| 30 | 98 | 1 | 30 | 98 | 1 | |
| 64 | 5 | 57 | 5 | |||
| 72 | 2:15 min | 72 | 2:15 min | |||
| 1 | 72 | 7 min | 1 | 72 | 7 min | |
| 1 | 4 | inf | 1 | 4 | inf |
Result
Expected Band: 4.711 Kb

l1:F1a without DMSO,l2:F1a with DMSO, l3:F16 without DMSO at an anealing temperature 64°C,l5:F16 with DMSO at an anealing temperature 64°C,l6:1b-57 (without DMSO), l7: 1b-57 (with DMSO),l9 2 loga ladder,l10 Mediprep A of BB,l11 Mediprep B of BBF1a with DMSO

l1:F1a without DMSO,l2:F1a with DMSO, l3:F16 without DMSO at an anealing temperature 64°C,l5:F16 with DMSO at an anealing temperature 64°C,l6:1b-57 (without DMSO), l7: 1b-57 (with DMSO),l9 2 loga ladder,l10 Mediprep A of BB,l11 Mediprep B of BBF1a with DMSO
PCR using DMSO at an annealing temperature od 57°C looks fine.
- => Fragment was cut and gel extracted.
Amplification of DelH G2
PCR Conditions G2.W12.A
| Reagent | DelH G2 | DelH G2 |
|---|---|---|
| Template | Fresh colony of plate (by DN) | Fresh colony of plate (by DN) |
| Primer fw 10 µM | 2 µl HM05 | 2 µl HM05 |
| Primer rev 10 µM | 2 µl HM08 | 2 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 8 µl | 7 µl |
| DMSO | - | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | |
| 72 | 3:30 min | |
| 18 | 98 | 1 |
| 67 | 5 | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 9.611 Kb
Expected band is visible.
- => Fragment G2 was amplified with DMSO. Band was cut and gel extracted.
PCR Conditions G2.W12.B
| Reagent | DelH G2 |
|---|---|
| Template | Fresh colony of plate (by DN) |
| Primer fw 10 µM | 2.5 µl HM05 |
| Primer rev 10 µM | 2.5 µl HM08 |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 17.5 µl |
| DMSO | 2.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | |
| 72 | 3:30 min | |
| 18 | 98 | 1 |
| 67 | 5 | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 9.611 Kb
Gel shows nice band at expected length.
- => G2 was also amplified in 50 µl and was cut and gel extracted.
Amplification of DelH G0
PCR Conditions G0.W12.A
| Reagent | DelH G0 |
|---|---|
| Template | 1 µl of glycerol stock |
| Expected length [Kb] | 18.521 |
| Primer fw 10 µM | 2.5 µl short2 |
| Primer rev 10 µM | 2.5 µl HM08 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 18.521 Kb
Gel shows expected band at ~18 Kb.
- => Fragment G0 was cut and gel isolated. Repeat PCR to increase yield.
Result
Three 20 µl reactions were performed to increase yield of fragment G0.
Expected band: 18.521 Kb
Gel shows expected band at ~18 Kb.
- => Fragment G0 was cut and gel isolated.
Amplification of DelH G1/2a
PCR Conditions G1/2a.W12.A
| Reagent | DelH G1/2a |
|---|---|
| Template | 1 µl of glycerol stock |
| Expected length [Kb] | 13.083 |
| Primer fw 10 µM | 2.5 µl short2 |
| Primer rev 10 µM | 2.5 µl HM06 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 13.083 Kb
Gel shows expected band at ~13 Kb.
- => Fragment G1/2a was cut and gel isolated.
Amplification of DelH G1b/2
PCR Conditions G1b/2.W12.A
| Reagent | DelH G1b/2 |
|---|---|
| Template | 1 µl of glycerol stock |
| Expected length [Kb] | 14.271 |
| Primer fw 10 µM | 2.5 µl HM03 |
| Primer rev 10 µM | 2.5 µl HM08 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 14.271 Kb
Gel shows expected band at ~14 Kb.
- => Fragment G1b/2 was cut and gel isolated.
Amplification of DelH G1b/2a
PCR Conditions G1b/2a.W12.A
| Reagent | DelH G1b/2a |
|---|---|
| Template | 1 µl of glycerol stock |
| Expected length [Kb] | 14.271 |
| Primer fw 10 µM | 2.5 µl HM03 |
| Primer rev 10 µM | 2.5 µl HM06 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 3:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 14.271 Kb
Gel shows multiple bands, it is not clear, which one is correct.
- => Optimize temperatures.
Amplification of DelH G2b
PCR Conditions G2b.W12.A
| Reagent | DelH G2b |
|---|---|
| Template | 1 µl of glycerol stock |
| Expected length [Kb] | 5 |
| Primer fw 10 µM | 2.5 µl HM07 |
| Primer rev 10 µM | 2.5 µl HM08 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:00 min | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
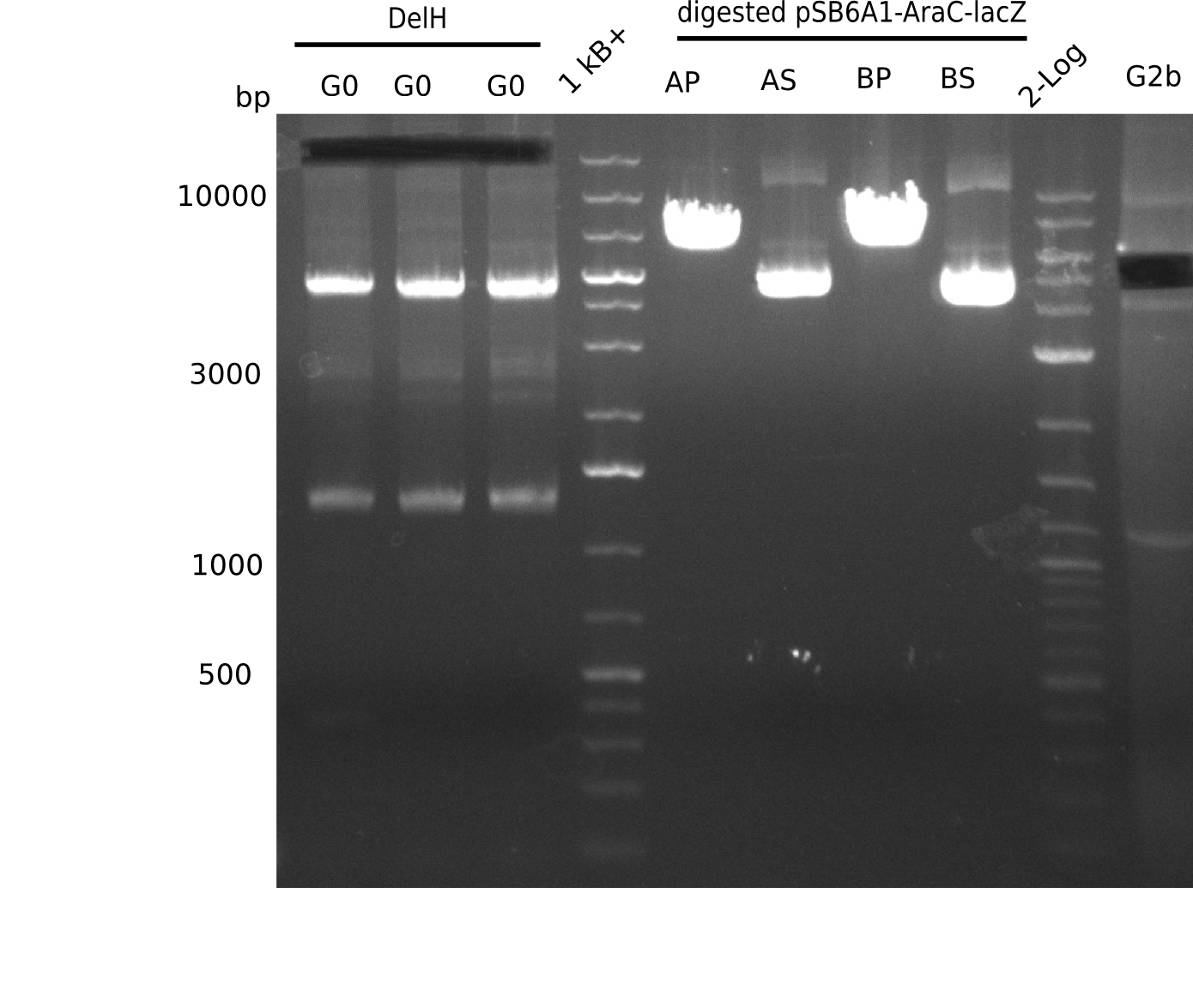
l1-3:DelH G0(18 Kb),l5:1 Kb+ ladder, l5:Midiprep A digested with PstI & BamHI,l6:Midiprep A digested with SalI & BamHI, l7:Midiprep B digested with PstI & BamHI,l8:Midiprep B digested with SalI & BamHI, l9: 2 log ladder, l10: fragment of DelH - G3b
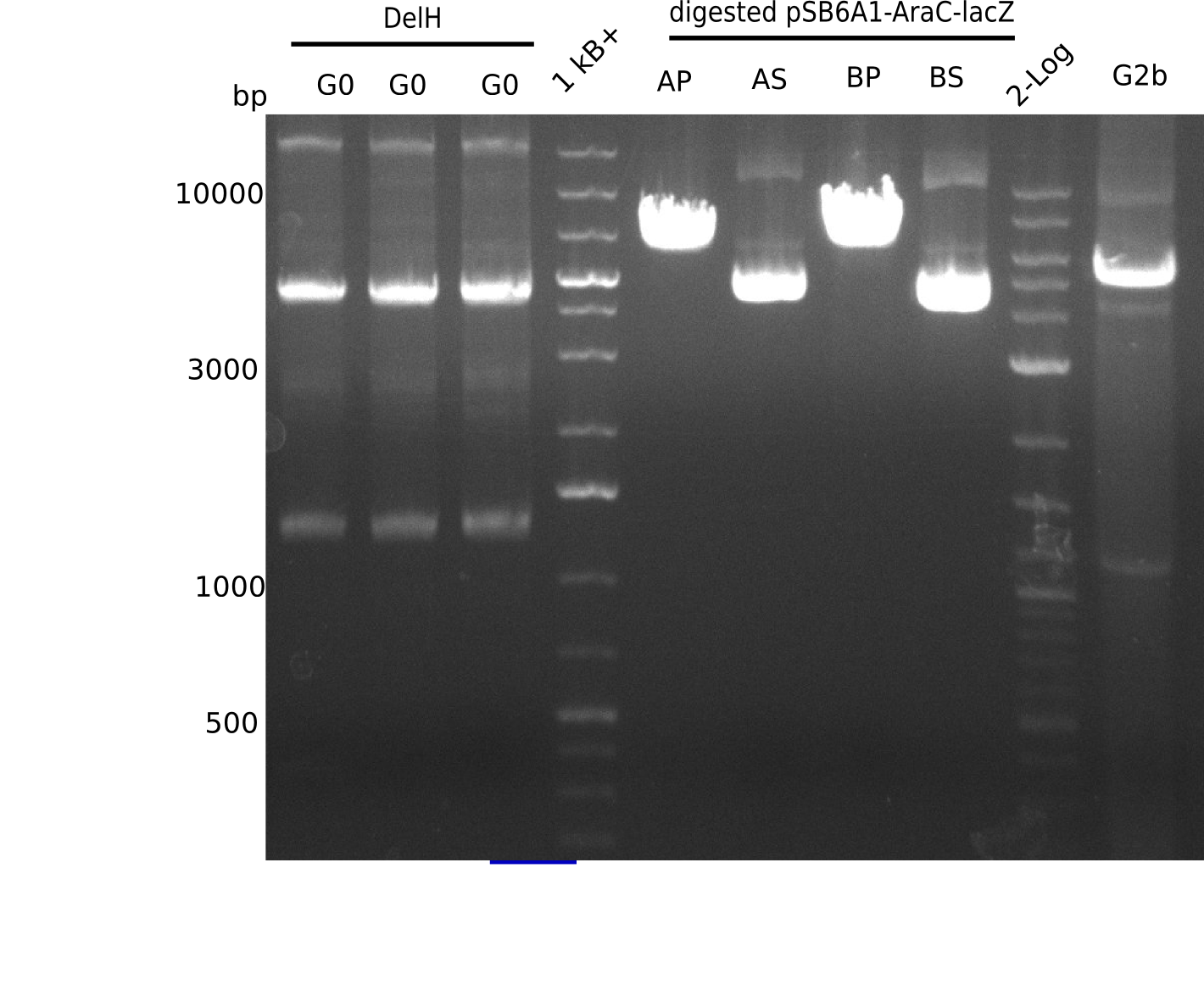
l1-3:DelH G0(18 Kb),l5:1 Kb+ ladder, l5:Midiprep A digested with PstI & BamHI,l6:Midiprep A digested with SalI & BamHI, l7:Midiprep B digested with PstI & BamHI,l8:Midiprep B digested with SalI & BamHI, l9: 2 log ladder, l10: fragment of DelH - G3b
Amplification of Backbone pSB6A1-AraC-lacZ
Test Restriction Digest of Midiprep
| Midiprep | A | A | B | B |
|---|---|---|---|---|
| Enzymes | BamHI & Pst | BamHI & SalI | BamHI & PstI | BamHI & SalI |
| Wanted fragments [Kb] | 3.317 & 4.071 | 3.794 & 3.594 | 3.317 & 4.071 | 3.794 & 3.594 |
| Fragments present | 1 band at ~6-7 Kb | 3 bands: one bright one at ~4-5 Kb and two smaller ones at ~6 Kb and ~10 Kb | 1 band at ~6-7 Kb | 3 bands: one bright one at ~4-5 Kb and two smaller ones at ~6 Kb and ~10 Kb |

l1-3:DelH G0(18 Kb),l5:1 Kb+ ladder, l5:Midiprep A digested with PstI & BamHI,l6:Midiprep A digested with SalI & BamHI, l7:Midiprep B digested with PstI & BamHI,l8:Midiprep B digested with SalI & BamHI, l9: 2 log ladder, l10: fragment of DelH - G3b

l1-3:DelH G0(18 Kb),l5:1 Kb+ ladder, l5:Midiprep A digested with PstI & BamHI,l6:Midiprep A digested with SalI & BamHI, l7:Midiprep B digested with PstI & BamHI,l8:Midiprep B digested with SalI & BamHI, l9: 2 log ladder, l10: fragment of DelH - G3b
- => Don't trust the BB, because between 3 and 4 Kb there is no band. Backbone will be send for sequencing
- => Repeat entire cloning again?
Amplification of Backbone pSB6A1-lacZ-mRFP
As discussed, we are going to use another backbone. It is already in the parts registry: pSB6A1 + BBa_J04450 from Spring distribution 2012 (plate 1, well K1).
Transformation in E. coli TOP10
- Chemical transformation of 3 µl of the plasmid
- Incubation on ice, 15 min
- Heat shock 42°C, 40 s
- Incubation on ice again
- Resuspended in 500 µl LB Amp for 30 min
- Centrifuged (120, 5,000 rpm), removal of supernatant
- Plated on LB Amp plate
- Incubation ON at 37°C
Amplification of 1
22-07 - 28-07-13
Amplification of Backbone pSB6A1-lacZ-mRFP
Miniprep
- 4 colonies were picked and inoculated in 4x 3 ml LB Amp
- Incubated ON at 37°C
- 4x 3 ml were distibuted in 5x 2 ml eppis and centrifuged
- Supernatant was discarded, cell pellet was used for miniprep
- Two minipreps were pooled into 1 column and 180ipreps on another one (every eluation was performed with 50 µl)
Result
- 5 µl were loaded on a gel
- => Gel was not succesful
- => Miniprep didn't work and has to be repeated
Glycerol Stock
Glycerol stocks were prepared from liquid culture not used for miniprep.
Repetiotion of Miniprep
4x 2 ml of Top10 pSB6A1-lacZ-mRFP were minipreped with the Qiagen kit.
Result
| Sample | Performed | Concentration (Nanovue) | P2 Buffer |
|---|---|---|---|
| pSB6A1-lacI-mRFP (1) | upstairs | 88 ng/µl | from upstairs |
| pSB6A1-lacI-mRFP (2) | iGEM lab | 62 ng/µl | new |
| pSB6A1-lacI-mRFP (3) | iGEM lab | 104 ng/µl | new |
| pSB6A1-lacI-mRFP (4) | iGEM lab | 60 ng/µl | old |
- Of each miniprep, 3 µl were loaded
At 3Kb and at 8 Kb, there are bands visible.
- => Minipreps are fine.
PCR Conditions BB.W13.A
| Reagent | Backbone | Backbone |
|---|---|---|
| Template | Miniprep A | Miniprep A |
| Primer fw 10 µM | HM11 | HM11 |
| Primer rev 10 µM | HM12 | HM12 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 7 µl | 7 µl |
| DMSO | - | - |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 (touchdown - 0,5) | 5 | |
| 72 | 1:15 min | |
| 18 | 98 | 1 |
| 67 | 5 | |
| 72 | 1:15 min | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
Specific expected band at 5 Kb
- => Fragment was cut and gel isolated. A: 22 ng/µl, B: 25 ng/µl
Generation of DelH Plasmid 26-07
Analysis of G0, G1 & G2b on Gel
| Fragment | Size | Amount used [µl] | Amount used [ng] (by view) | Left for Gibson [µl] = [ng] | Concentration [ng/µl] |
|---|---|---|---|---|---|
| G0 | 18 Kb | 8 | 5 | 52 = 35.5 | 0.68 |
| G1 | 14 Kb | 8 | 10 | 12 = 15 | 1.25 |
| G2b | 5 Kb | 8 | 10 | 12 = 15 | 1.25 |
Overview on Fragments
| Fragment | Concentration [ng/µl] |
|---|---|
| G0 | 0.68 |
| G1 | 1.25 |
| G2b | 1.25 |
| BB A | 22 |
| BB B | 25 |
Gibson Assembly
| Assembly # | Fragments | Amount used [µl] |
|---|---|---|
| 1 |
|
|
| 2 |
|
|
| 3 |
|
|
- Incubated 1 hour at 50°C
- Stored ON at -20°C
Purification of Gibson Mix
- Purification performed with the long range gel extraction kit (because of 23 Kb size) of the entire sample (20 µl).
- Performed the purification by calculating with 100 mg DNA (gel- step1)
- Washing step with QX1 was not performed, because it is only to remove the residual agarose and we don't want to loose any DNA.
Electroporation
- Electroporation into freshly prepared electrocompetent cell following the protocol
- Streaked on LB Amp plates and incubated ON at 37°C
Result
There were no colonies on any of the plates. Ginson assembly and electroporation have to be repeated.
Generation of DelH Plasmid 28-07
NEB Protocol for Gibson Assembly
- Optimized cloning efficiency is 50–100 ng of vectors with 2–3 fold of excess inserts. Use 5 times more of inserts if size is less than 200 bps. Optimized cloning efficiency is 50-100 ng of vectors with 2-3 fold of excess inserts. Use 5 times more of inserts if size is less than 200 bps.
- Electrocompetent Cells Transformation Protocol:
- Thaw electrocompetent cells on ice.
- Transfer 50 μl of electrocompetent cells to a pre-chilled electroporation cuvette with 1 mM gap.
- Dilute assembled products 3-fold with H2O prior electroporation. This can be achieved by mixing 5 μl of assembled products with 10 μl of H2O. Add 1 μ l of the diluted assembly product to electrocompetent cells.Mix gently by pipetting up and down or flicking the tube 4–5 times. Do not vortex. Place the mixture on ice for 30 minutes. Do not mix.
- Mix gently by pipetting up.
- Once DNA is added to the cells, electroporation can be carried out immediately. It is not necessary to incubate DNA with cells.Add 950 μl of room temperature SOC media* to tubes.
- Add 950 μl of room temperature SOC media to the cuvette immediately after electroporation.
- Place the tube at 37°C for 60 minutes. Shake vigorously (250 rpm) or rotate.
- Warm selection plates to 37°C.
- Spread 100 μl of the cells onto the plates.
- Incubate overnight at 37°C.
Gibson Assembly
| Fragments | Concentraion [ng/µl] | Amount [µl] |
|---|---|---|
| G0 | 0.68 | 9 |
| BB (8.8) | 22 | 1 |
| Gibson Master Mix | 2x | 10 |
- Incubated 1 h at 37 °C in thermocycler
- 5 µl of mix added to 10 µl ddH2O and stored at -20°C
- 10 µl stored for isoprop purification
Result
Remaining 5 µl were checked on a gel:
Expected band: 23 Kb
No product visible of correct size. There is a light band at ~1.5 Kb.
- => Most probably too little DNA loaded.
Electroporation
- Electroporation into freshly prepared electrocompetent cell following the protocol
- Streaked on LB Amp plates and incubated ON at 37°C
Result
There were few colonies on the plates. Red colonies were picked and screenied by colony-PCR, but none of them was positive. The Gibson assembly has to be repeated.
Preparation of Electrocompetent E.coli DH10ß
According to the protocol
Amplification of Backbone pSB6A1-lacZ-mRFP
Test Restriction Digest
| Fragment | DNA [µl] | H2O [µl] | Enzymes [µl] | Buffer 3.1 [µl] |
|---|---|---|---|---|
| BB A (22 ng/µl) | 8 = 200 ng | 9 | NotI: 1 | 2 |
- Incubated 1.5 h at 37 °C
Result
Expected bands: 4 Kb & 1 Kb
Gel shows shows expeted bands at 4 Kb and 1 Kb.
- => Backbone is fine.
Amplification of 1
29-07 - 04-08-13
Generation of DelH Plasmid 28-07
Purification of Gibson Mix
10 µl of the Gibson mix were isopropanol purified as described in the protocol.
5 µl of the Gibson reaction were mixed with 10 µl ddH2O.
Electroporation
- Electroporation into freshly prepared electrocompetent cell following the protocol.
| E1 | E2 | E3 |
|---|---|---|
| 5 µl of Gibson + 10 µl H2O | 10 µl of isoprop purified Gibson assembly | no DNA |
- Grow 0.5 h with 400 µl SOC at 37°C
- Add 9 ml LB Amp and incubate 3 h at 37°C
- Incubate ON at 37°C on LB Amp plate as well as LB (no Amp) for control purposes
Result
There were few colonies on the plates. Red colonies were picked and screenied by colony-PCR, but none of them was positive. The Gibson assembly has to be repeated.
Generation of DelH Plasmid 01-08
Gibson Assembly
| Fragment | Concentraion | Amount used [µl] (2 vials) | Amount used [µl] |
|---|---|---|---|
| G0 | 0,68 ng/µl | 9 | 6 |
| BB (8.8) | 22 ng/µl | 1 | 4 |
| Gibson Master Mix | 2x | 10 | 10 |
- Incubated 1 h at 50 °C in thermocycler
Electroporation
| Electroporation # | Mix # | Amount of Mix [µl] | ddH2O [µl] | Isopropanol | Amount electroporated [µl] |
|---|---|---|---|---|---|
| 1 | 1 | 10 | 20 | - | 1 |
| 2 | 1 | 10 | 20 | - | 14 |
| 3 | 1 | 30 | - | yes | 20 |
| 4 | 2 | 5 | 10 | - | 1 |
| 5 | 2 | 5 | 10 | - | 29 |
| 6 | 2 | 15 | - | yes | 20 |
| 7 | - | - | 10 | - | 10 |
- Plated on one LB agar plate each
- Stored ON at 37°C
Colony-PCR Conditions CP.W14.A
| Reagent | Electroporation 1 | Electroporation 2 | Electroporation 3 | Electroporation 4 | Electroporation 5 | Electroporation 6 |
|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 1 | 2 | 3 | 4 | 5 | 6 |
| Template | 5 colonies of E1 | 5 colonies of E2 | 5 colonies of E3 | 5 colonies of E4 | 5 colonies of E5 | 5 colonies of E6 |
| Primer fw 10 µM | 1 µl VF2 | 1 µl VF2 | 1 µl VF2 | 1 µl VF2 | 1 µl VF2 | 1 µl VF2 |
| Primer rev 10 µM | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 8 µl | 8 µl | 8 µl | 8 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Gel does not show any band.
- => None of the analyzed colonies is positive.
Colony-PCR Conditions CP.W14.B
Picked 20 colonies per plate and 5 colonies per PCR tube.
| Reagent | Electr. 1 | Electr. 1 | Electr. 1 | Electr. 2 | Electr. 2 | Electr. 2 | Electr. 2 |
|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 1a | 1b | 1c | 2a | 2b | 2c | 2d |
| Template | 5 colonies of E1 | 5 colonies of E1 | 5 colonies of E1 | 5 colonies of E2 | 5 colonies of E2 | 5 colonies of E2 | 5 colonies of E2 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Reagent | Electr. 3 | Electr. 3 | Electr. 3 | Electr. 3 | Electr. 4 | Electr. 4 | Electr. 4 | Electr. 4 |
|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 3a | 3b | 3c | 3d | 4a | 4b | 4c | 4d |
| Template | 5 colonies of E3 | 5 colonies of E3 | 5 colonies of E3 | 5 colonies of E3 | 5 colonies of E4 | 5 colonies of E4 | 5 colonies of E4 | 5 colonies of E4 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6µl | 6 µl | 6 µl |
| Reagent | Electr. 5 | Electr. 5 | Electr. 5 | Electr. 5 | Electr. 6 | Electr. 6 | Electr. 6 | Electr. 6 |
|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 5a | 5b | 5c | 5d | 6a | 6b | 6c | 6d |
| Template | 5 colonies of E5 | 5 colonies of E5 | 5 colonies of E5 | 5 colonies of E5 | 5 colonies of E6 | 5 colonies of E6 | 5 colonies of E6 | 5 colonies of E6 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp

l1: 2log, l2-5: 20 picked colonies from plate 4: every PCR tube has 5 colonies, l6-9:20 picked colonies from plate 5every PCR tube has 5 colonies,l10-14:20 picked colonies from plate 6 every PCR tube has 5 colonies
bands on lanes 4a, 4b, 6a, 6b, 6d at the expected 663 bp, so the next step is making PCRs of 1 colony per PCR of these probes

l1-3:3 picked colonies from plate 1, every PCR tube has 5 colonies, l4-7:4 picked colonies from plate 2, every PCR tube has 5 colonies,l8: 2log,l9-13:4 picked colonies from plate 3 every PCR tube has 5 colonies
bands on lanes 3b, 3c, 3d, 4a, 4b, 6a, 6b, 6d at the expected 663 bp, so the next step is making PCRs of 1 colony per PCR of these probes
Gel shows bands for b, 3c, 3d, 4a, 4b, 6a, 6b, 6d at the expected 663 bp.
- => Make single colony-PCRs.
Colony-PCR Conditions CP.W14.C
1 µl of colony ON culture picked
| Reagent | 3c | 3c | 3c | 3c | 3c |
|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 |
| Named | 3c 1 | 3c 2 | 3c 3 | 3c 4 | 3c 5 |
| Template (1 µl) | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5µl | 5 µl | 5 µl |
| Reagent | 4a | 4a | 4a | 4a | 4a |
|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 |
| Named | 4a 1 | 4a 2 | 4a 3 | 4a 4 | 4a 5 |
| Template (1 µl) | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
| Reagent | 6b | 6b | 6b | 6b | 6b |
|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 |
| Named | 6b 1 | 6b 2 | 6b 3 | 6b 4 | 6b 5 |
| Template (1 µl) | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5µl | 5 µl | 5 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
- => No band is visible. There should be a band at at least one of each sample (for example 6B), because in the previous PCR we observed a band.
Because of the explained reason above we incubated 40 colonies seperately which might be positive ON at 37°C in 200 µl LB Amp. 200 µl of the ON cultire was mixed with 1.8 ml LB Amp for five colonies of each probe: 3b, 3c, 3d, 4a, 4b, 6a, 6b, 6d.
Amplification of 1
05-08 - 13-08-13
Generation of DelH Plasmid 01-08
Colony-PCR Conditions CP.W15.A
From colonies inoculated on 04-08
| Reagent | 3c | 3c | 3c | 3c | 3c |
|---|---|---|---|---|---|
| Expected length [kb] | 663 | 663 | 663 | 663 | 663 |
| Named | 3c 1 | 3c 2 | 3c 3 | 3c 4 | 3c 5 |
| Template (1 µl) | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5µl | 5 µl | 5 µl |
| Reagent | 4a | 4a | 4a | 4a | 4a |
|---|---|---|---|---|---|
| Expected length [kb] | 663 | 663 | 663 | 663 | 663 |
| Named | 4a 1 | 4a 2 | 4a 3 | 4a 4 | 4a 5 |
| Template (1 µl) | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
| Reagent | 6b | 6b | 6b | 6b | 6b |
|---|---|---|---|---|---|
| Expected length [kb] | 663 | 663 | 663 | 663 | 663 |
| Named | 6b 1 | 6b 2 | 6b 3 | 6b 4 | 6b 5 |
| Template (1 µl) | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
None of the gels shows expected band.
- => None of the colonies is positive.
Miniprep
Of colonies 6b(1)-6b(5), 6a(1), 6a(2), 3c(3), a miniprep was performed.
| Sample | Concentration [ng/µl] | µl avaiblable |
|---|---|---|
| 6b(1) | 33.8 | 18 |
| 6b(2) | 33.4 | 18 |
| 6b(3) | 36.9 | 18 |
| 6b(4) | 59.6 | 18 |
| 6b(5) | 18 | 18 |
| 6a(1) | 60 | 18 |
| 6a(2) | 62.3 | 18 |
| 3c(3) | 40.9 | 18 |
Test Restriction Digest
| Sample | Miniprep [µl] | CutSmart Buffer [µl] | NotI-HF [µl] | H2O [µl] | Total [µl] |
|---|---|---|---|---|---|
| 6b(1) | 15 | 2 | 1 | 3 | 20 |
| 6b(2) | 15 | 2 | 1 | 3 | 20 |
| 6b(3) | 15 | 2 | 1 | 3 | 20 |
| 6b(4) | 10 | 2 | 1 | 8 | 20 |
| 6b(5) | 15 | 2 | 1 | 3 | 20 |
| 6a(1) | 10 | 2 | 1 | 8 | 20 |
| 6a(2) | 10 | 2 | 1 | 8 | 20 |
| 3c(3) | 15 | 2 | 1 | 3 | 20 |
Result
Expected bands: 11,079 bp, 6,201 bp, 3,998 bp, 2,371 bp
| Expected bands | Present bands |
|---|---|
| 11.079 bp | ? |
| 6.201 bp | no |
| 3.998 bp | yes |
| 2.371 bp | no |
in silico PCR of BB shows the following result:
- 2 fragments generated:
- 1: 3.998 bp - From NotI[9] To NotI[4007]
- 2: 1.093 bp - From NotI[4007] To NotI[9]
But an additional band between 1-2 Kb is present!
- => Talk to Advisors, because it isn't what we expected. What could it be?
Possible next steps:
- a) Wait for the new strain of D. acidovorans SHP1
- b) Perform a re-PCR of minipreps (one with BB and one with DelH G0 fragment)
Generation of Backbone pSB6A1-lacI-mRFP
Restriction Digest with DpnI
After consulting the advisors, we decided not to continue the strategy with the old construct and wait with DelH until the new strain of D. acidovorans arrives. Until then, we digest the backbone (which is used also in following experiments) with DpnI to get rid of any remaining template.
| Reagent | Amount [µl] |
|---|---|
| Gel extracted lineralized by PCR (template=10.4 Kb) | 17 |
| DpnI | 1 |
| CutSmart buffer | 2 |
| Total | 20 |
- 1 h incubation at 37°C
- 20 min inactivation of enzyme by heat shock at 80°C for 20 min
Purification
- As template, the PCR of 10,4 PCR, which was digested with DpnI on 06-08, was used.
- Purification was performed with nucleotide removal kit (wrong column was used, but there was some yield at the nanodrop)
=> Again, checking on gel (2 µl + 8 µl 1xloading dye)
- => Concentration at nanodrop was 27.2 ng/µl
Amplification of DelH G0
PCR Conditions G0.W15.A
| Reagent | Amount [µl] |
|---|---|
| Expected length [Kb] | 18.521 |
| Named | G0 |
| Template | 1 µl New strain culture SHP1 (resuspended pellet) |
| Primer fw 10 µM | 1 µl short2 |
| Primer rev 10 µM | 1 µl HM08 |
| Phusion Flash Ready Mix | 10 |
| ddH2O | 5 |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18.521 Kb
Gel does not show the expected band.
- => Further improve PCR conditions.
PCR Conditions G0.W15.B
| Reagent | Amount [µl] |
|---|---|
| Expected length [Kb] | 18.521 |
| Named | G0 |
| Template | Picked colony |
| Primer fw 10 µM | 1 µl short2 |
| Primer rev 10 µM | 1 µl HM08 |
| Phusion Flash Ready Mix | 10 |
| ddH2O | 5 |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
- => Becaus it worked well we amplified 3x G0 with the same conditions.
PCR Conditions G0.W15.C
3x 20 µl PCR reactions were prepared
| Reagent | Amount [µl] |
|---|---|
| Expected length [Kb] | 18.521 |
| Named | G0 |
| Template | Picked colony |
| Primer fw 10 µM | 1 µl short2 |
| Primer rev 10 µM | 1 µl HM08 |
| Phusion Flash Ready Mix | 10 |
| ddH2O | 5 |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb
Gel shows the expected bands.
- => Fragments were cut and gel extracted.
Amplification of DelH G1/2a
PCR Conditions G1/2a.W15.A
| Reagent | Amount [µl] | Amount [µl] |
|---|---|---|
| Expected length [Kb] | 13.083 | 13.083 |
| Named | G1/2a | G1/2a |
| Template | Picked colony | Picked colony |
| Primer fw 10 µM | 1 µl short2 | 1 µl short2 |
| Primer rev 10 µM | 1 µl HM06 | 1 µl HM06 |
| Phusion Flash Ready Mix | 10 | 10 |
| ddH2O | 5 | 4 |
| DMSO | 1 | 0 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Gel shows the expected bands.
- => Fragments were cut and gel extracted.
PCR Conditions G1/2a.W15.B
| Reagent | Amount [µl] |
|---|---|
| Expected length [Kb] | 13.083 |
| Named | G1/2a |
| Template | Picked colony |
| Primer fw 10 µM | 1 µl short2 |
| Primer rev 10 µM | 1 µl HM06 |
| Phusion Flash Ready Mix | 10 |
| ddH2O | 5 |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 13 Kb
Gel shows the expected bands.
- => Fragments were cut and gel extracted.
Amplification of DelH G2b
PCR Conditions G2b.W15.A
| Reagent | Amount [µl] | Amount [µl] |
|---|---|---|
| Expected length [Kb] | 5.472 | 5.472 |
| Named | G2b | G2b |
| Template | Picked colony | Picked colony |
| Primer fw 10 µM | 1 µl HM07 | 1 µl HM07 |
| Primer rev 10 µM | 1 µl HM08 | 1 µl HM08 |
| Phusion Flash Ready Mix | 10 | 10 |
| ddH2O | 5 | 4 |
| DMSO | 1 | 0 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Summary of Gel Extracted Fragments
Concentrations (by nanodrop) are summarized in the following table and gel picture.
| Fragment | Concentration [ng/µl] |
|---|---|
| G0 | 3.5 |
| G1/2a | 4.9 |
| G2b | 18.6 |
Generation of DelH Plasmid 09-08
Gibson Assembly
We decided two assemble 2 different constructs and one control (only the backbone).
| Construct | G0 [µl] | G1/2a [µl] | G2b [µl] | BB [µl] | 2x Gibson Master Mix [µl] | Final amount [µl] |
|---|---|---|---|---|---|---|
| 1 | 9 | 0 | 0 | 1 | 10 | 20 |
| 2 | 0 | 8 | 1 | 1 | 10 | 20 |
| Control (C) | 0 | 0 | 0 | 1 (+9 ddH2O ) | 10 | 20 |
- Incubated 1 h at 50°C
Electroporation
- Of each Gibson assembly, 5 µl are diluted in 10 µl ddH2O
- The rest of the Gibson assembly is stored at -20°C
Conoly-PCR Conditions CP.W15.A
Picked 8 colonies from plates with 10µl and 100 µl construct 1, and plate with 100 µl of construct 2. 1 additional colony was picked from plate with 10 µl of construct 2 (because there were no more cells) and 1 colony of the control(100µl), and one colony of the control (100µl) with new electrocompetent cells.
- In the following table construct 1 is named C1, and construct 2 = C2 and Control = Co
| Reagent | C1 (10 µl) | C1 (10 µl) | C1 (10 µl) | C1 (10 µl) | C1 (10 µl) | C1 (10 µl) | C1 (10 µl) | C1 (10 µl) |
|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Reagent | C1 (100 µl) | C1 (100 µl) | C1 (100 µl) | C1 (100 µl) | C1 (100 µl) | C1 (100 µl) | C1 (100 µl) | C1 (100 µl) |
|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6µl | 6 µl | 6 µl | 6 µl |
| Reagent | C2 (10 µl) | C2 (10 µl) | C2 (10 µl) | C2 (10 µl) | C2 (10 µl) | C2 (10 µl) | C2 (10 µl) | C2 (10 µl)) |
|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Reagent | C2 (10 µl) | C2 (100 µl) | Co (100 µl) | Co(100 µl) |
|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | no product | no product |
| Named | 24 | 25 | 26 | 27 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Gel shows no banda at 663 bp. Instead, they show an unexpected band at ~6 Kb.
- => None of the colonies is positive.
Conoly-PCR Conditions CP.W15.B
Picked 20 colonies from plates with 10 µl and 100 µl construct 1 and construct 2 (80 colonies). 4 colonies were picked from the control (10 µl) and control (100 µl).
- In the following table construct 1 is named C1, and construct 2 = C2 and Control = Co
| Reagent | C1 (10 µl) | C1 (10 µl) | C1 (10 µl) | C1 (10 µl) | C1 (100 µl) | C1 (100 µl) | C1 (100 µl) | C1 (100 µl) |
|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | A | B | C | D | E | F | G | H |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Reagent | C2 (10µl) | C2 (10µl) | C2 (10µl) | C2 (10µl) | C2 (100µl) | C2 (100µl) | C2 (100µl) | C2 (100µl) |
|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | I | J | K | L | M | N | O | P |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Reagent | Co (10µl) | Co (10µl) | Co (10µl) | Co (10µl) | Co(100µl) | Co(100µl) | Co(100µl) | Co(100µl) |
|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
- => New Gibson Assembly will be performed
Gel shows no banda at 663 bp.
- => Neither the colonies A-P show the expected band, nor the control shows any band.
Generation of DelH Plasmid 11-08
Gibson Assembly
Because of the past results, we assume that the colonies can grow because the backbone religates. We also found out that the first overhang of the backbone has some homologity with a sequence at the end and we can assume that with Gibson, it primes itself.
So in the following step we will use less backbone. We prepared an 1:10 dilution of the backbone construct.
| Reagent | Concentration [ng/µl] | Amount [µl] Mix A | Amount [µl] Mix B | Amount [µl] Mix C |
|---|---|---|---|---|
| G0 | 3.5 | 9 | 0 | 0 |
| BB | 1.8 | 1 | 1 | 1 |
| Gibson Assembly Master Mix | 2x | 10 | 10 | 0 |
The time constant for the electroporation was unexpectedly low, hopefully high enough...
Amplification of 1
12-08 - 18-08-13
Increasing Yield of DelH Fragments
PCR Conditions G0.G1/2a.W16.A
| Reagent | G0 | G0 | G0 | G1/2a | G1/2a |
|---|---|---|---|---|---|
| Expected length [Kb] | 18 | 18 | 18 | 13.083 | 13.083 |
| Named | G0 | G0 | G0 | 1 | 1 |
| Template | Picked colony | Picked colony | Picked colony | Picked colony | Picked colony |
| Primer fw 10 µM | 1 µl short2 | 1 µl short2 | 1 µl short2 | 1 µl HM07 | 1 µl HM07 |
| Primer rev 10 µM | 1 µl HM08 | 1 µl HM08 | 1 µl HM08 | 1 µl HM06 | 1 µl HM06 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
| DMSO | 1 µl | 1 µl | 1 µl | 1 µl | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start used
Result
Expected bands: 18 Kb (G0), 13 Kb (G1/2a)
Gel shows expected bands.
- => They were cut and gel isolated.
Generation of Plasmid DelH 11-08
Colony-PCR CP.W16.A
| Reagent | Cells + 1 µl Gibson Assembly | Cells + 14 µl Gibson Assembly | Cells + 14 µl Gibson Assembly (ONLY Backbone) | Cells + 14 µl Gibson Assembly (ONLY Backbone) |
|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | - | - |
| Named | D1 | D14 | B14(1) | B14(2) |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
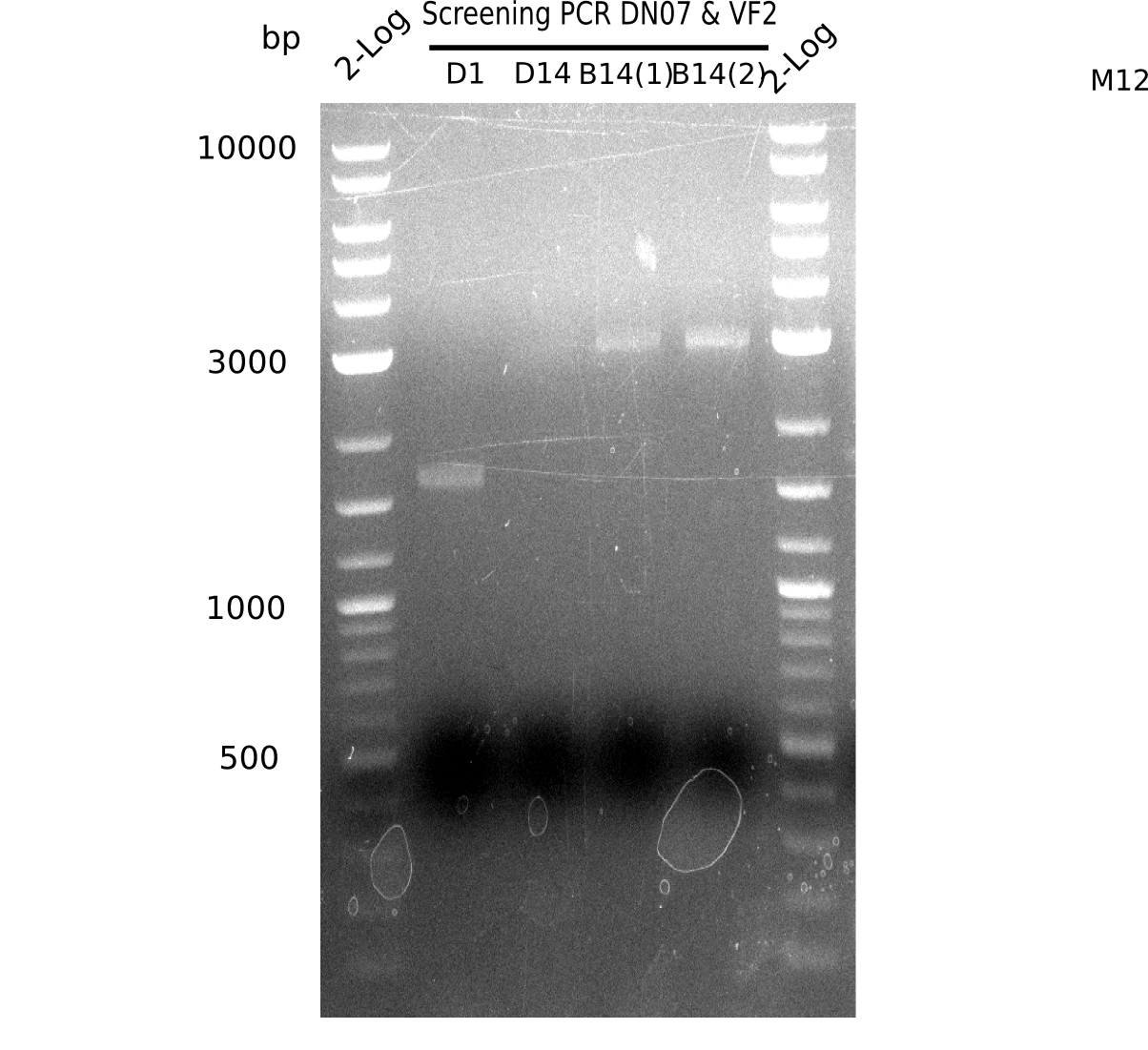
l1: 2log ladder, l2: colony PCR of 1 µl Gibson transformed,l3: colony PCR of 14 µl Gibson transformed,l4-5: 2 colony PCRs of BB - 14 µl Gibson transformed, l6: 2log ladder
none of the colonies is positive, of course the BB shows not the expected band of 663 bp (for screening). The first colony shows a band at ~1.7 kb.
None of the colonies is positive, of course the BB shows not the expected band of 663 bp (for screening). The first colony shows a band at ~1.7 kb.
- => None of the colonies harbours DelH plasmid.
Colony-PCR CP.W16.B
| Reagent | Cells + 10 µl Gibson Assembly pink | Cells + 10 µl Gibson Assembly pink | Cells + 10 µl Gibson Assembly white | Cells + 10 µl Gibson Assembly white |
|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 |
| Named | A | B | C | D |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5 °C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Testing of Screening Primers
To test the reverse screening primer DN07 we amplify with the DreamTaqPolymerase and DN11 primer. As template, we use the complete DelH (18 Kb) and the first 3/4 of DelH (14 Kb).
| Reagent | 1 µl G0 of 23-07 | 1 µl G1/2a of 23-07 |
|---|---|---|
| Expected length [bp] | 323 | 323 |
| Named | 0 | 1 |
| Primer fw 10 µM | 2 µl DN11 | 2 µl DN11 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 30 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 30 | |
| 1 | 12 | inf |
Result
Expected length= 323 bp for positive clones
Gel shows expected bands.
- => Both PCRs were positive. Proof of functionality of DN07 screening primer and that both fragments are the expected DelH fragments.
Amplification of DelH G0
Gradient PCR Conditions G0.W16.B
To improve the PCR of the fragment with the length of 18 Kb, we run a gradient PCR between 68°C and 65°C.
| Reagent | G0 | G0 | G0 | G0 |
|---|---|---|---|---|
| Expected length [Kb] | 18 | 18 | 18 | 18 |
| Named | G0 1 | G0 2 | G0 3 | G0 4 |
| Template | Picked colony | Picked colony | Picked colony | Picked colony |
| Primer fw 10 µM | 2 µl short2 | 2 µl short2 | 2 µl short2 | 2 µl short2 |
| Primer rev 10 µM | 2 µl HM08 | 2 µl HM08 | 2 µl HM08 | 2 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5 µl | 5 µl |
| DMSO | 1 µl | 1 µl | 1 µl | 1 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 - 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb
Gel does not show expected band.
- => Further optimize PCR conditions.
PCR Conditions G0.W16.C
Because the amplification of DelH GO didn't work yesterday, we try it again with one picked colony and 1 µl of a glycerol stock at a constant annealing temperature of 65°C. 65°C was the temperature on which it worked the last time.
| Reagent | G0 | G0 |
|---|---|---|
| Expected length [Kb] | 18 | 18 |
| Named | G0 1 | G0 2 |
| Template | Picked colony SPH1 | 1 µl of glycerol stock SPH1 |
| Primer fw 10 µM | 2 µl short2 | 2 µl short2 |
| Primer rev 10 µM | 2 µl HM08 | 2 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 5 µl | 4 µl |
| DMSO | 1 µl | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb
Gel shows expected band.
- => It was cut and gel extracted. Also, we will run again 3 samples with the same conditions.
PCR Conditions G0.W16.D
| Reagent | G0 | G0 | G0 |
|---|---|---|---|
| Expected length [Kb] | 18 | 18 | 18 |
| Named | G0 1 | G0 2 | G0 2 |
| Template | 1 µl of glycerol stock SPH1 | 1 µl of glycerol stock SPH1 | 1 µl of glycerol stock SPH1 |
| Primer fw 10 µM | 2 µl short2 | 2 µl short2 | 2 µl short2 |
| Primer rev 10 µM | 2 µl HM08 | 2 µl HM08 | 2 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl |
| ddH2O | 4 µl | 4 µl | 4 µl |
| DMSO | 1 µl | 1 µl | 1 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb
Gel shows expected fragments.
- => They were cut and gel extracted.
Amplification of DelH G1/2a
Gradient PCR Conditions G1/2a.W16.B
To improve the PCR of the fragment with the length of 18 Kb, we run a gradient PCR between 68°C and 65°C.
| Reagent | G1/2a | G1/2a | G1/2a | G1/2a |
|---|---|---|---|---|
| Expected length [Kb] | 13.083 | 13.083 | 13.083 | 13.083 |
| Named | 1 1 | 1 2 | 1 3 | 1 4 |
| Template | Picked colony | Picked colony | Picked colony | Picked colony |
| Primer fw 10 µM | 2 µl short2 | 2 µl short2 | 2 µl short2 | 2 µl short2 |
| Primer rev 10 µM | 2 µl HM06 | 2 µl HM06 | 2 µl HM06 | 2 µl HM06 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5 µl | 5 µl |
| DMSO | 1 µl | 1 µl | 1 µl | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 - 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 13 Kb
Gel shows expected band of G1/2a, but also additional uspecific bands.
- => Fragment was cut and gel extracted.
Generation of Plasmid DelH 17-08
Electroporation
Remaining Gibson mix from 01.08 was electroporated.
Colony-PCR CP.W16.B
5 colony per PCR tube (but in different LB Amp Eppis)
| Reagent | Electroporation from 17-08 10 µl | Electroporation from 17-08 100 µl |
|---|---|---|
| Expected length [bp] | 663 | 663 |
| Named | 1 - 2 | 3 - 12 |
| Template | 2 colonies of 10 µl | 10 colonies of 100 µl |
| Primer fw 10 µM | 1 µl VF2 | 1 µl VF2 |
| Primer rev 10 µM | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl |
| ddH2O | 8 µl | 8 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
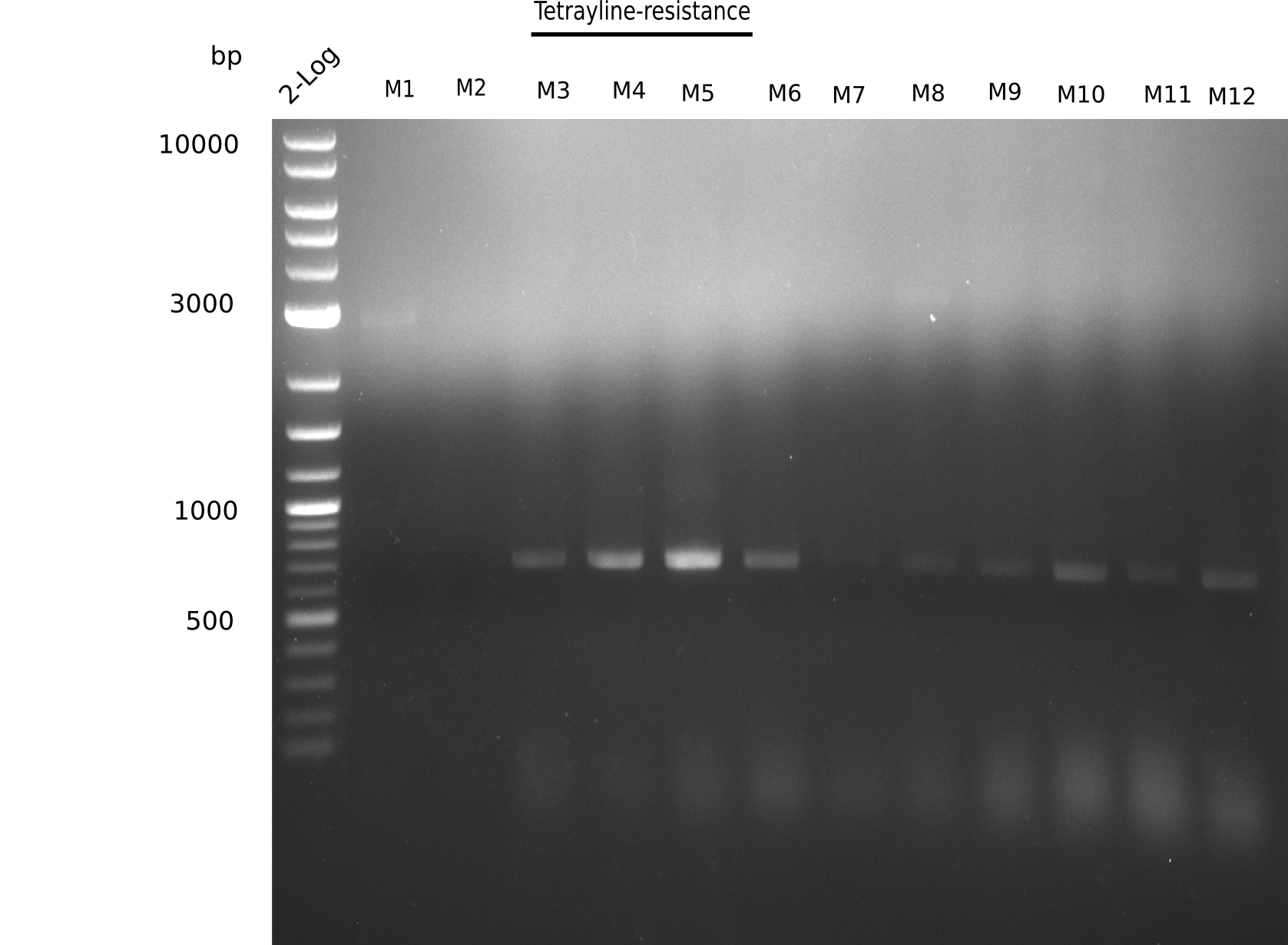
l1: 2log ladder, l2: 1, l3: 2, l4: 3, l5: 4, l6: 5, l7: 6, l8: 7, l9: 8, l10: 9, l11: 10, l12: 11, l13: 12
l4-13 show expected band = positive?
Majority of colonies show expected band.
- => Grow ON, perform mini prep and test digests as well as PCRs of fragments from isolated DNA.
Amplification of 1
19-08 - 25-08-13
Summary of Amplified DelH Fragments
| Fragment | Concentration [ng/µl] |
|---|---|
| G0 | 6.4 |
| G1/2a | 8.5 |
Rather low yield. So we switched the strategy and try to increase the template of DelH by amplifying it in smaller and more fragments. Therefore, we used the following primers: FS64-FS71, HM08, DN07
Amplification of DelH G0
Re-PCR Conditions G0.W17.A
| Reagent | G0 |
|---|---|
| Expected length [Kb] | 18 |
| Named | G0 1 |
| Template | 1 µl G0 fragment (c= 6.4 ng/µl) |
| Primer fw 10 µM | 2 µl short2 |
| Primer rev 10 µM | 2 µl HM08 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb

l1: BB amplified with HM16 by 2-step PCR, l2: BB amplified with HM17by 2-step PCR, l3: BB amplified with HM16, l4: BB amplified with HM17, l5: DelH-G0
l5: G0 was not amplified => not our fragment or without DMSO it is not so effective
Gel does not show expected band.
- => G0 was not amplified. Possibly change Mg2+ concentration.
PCR Conditions G0.W17.B
| Reagent | G0 | G0 | G0 | G0 |
|---|---|---|---|---|
| Expected length [Kb] | 18 | 18 | 18 | 18 |
| Named | G0 0.5 | G0 1 | G0 2 | G0 N |
| Template | 1 µl glycerol stock D. acidovorans | 1 µl glycerol stock D. acidovorans | 1 µl glycerol stock D. acidovorans | 1 µl glycerol stock D. acidovorans |
| Primer fw 10 µM | 2 µl short2 | 2 µl short2 | 2 µl short2 | 2 µl short2 |
| Primer rev 10 µM | 2 µl HM08 | 2 µl HM08 | 2 µl HM08 | 2 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 4.5 µl | 4 µl | 3.5 µl | 4 µl |
| DMSO | 1 | 1 | 1 | 1 |
| MgSO4 | 0.5 µl | 1 µl | 1.5 µl | - |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb
Fragments were amplified, but yield not significantly increased.
- => DMSO is important, but MgSO4 is not improve the PCR reaction.
Characterization of DelH Plasmid pHM03 17-08
- For the miniprep, the cells were centrifuged at 13,000 rpm for 15 min
- The cell pellet was resuspended in 200 µl P1 + 400 µl P2 and incubated for 120
- Afterwards, 300 µl S3 were added and transferred into a new 2 ml eppi and centrifuged 20 min at 13,000 rpm
- 1 ml isopropanol was added and the normal isoprop-ethanol-preipitation
- It was resuspended in 35 µl ddH2O
Result
| Miniprep of colony | Concentration [ng/µl] |
|---|---|
| 3 | 1,868.5 |
| 4 | 1,688.1 |
| 5 | 1,069.2 |
| 6 | 1,778.6 |
| 7 | 2,105.7 |
| 8 | 1,219.7 |
| 9 | 1,083.1 |
| 10 | 1,031.7 |
| 11 | 2,175.3 |
| 12 | 1,449.4 |
PCR Conditions CP.W17.A
M3 = Miniprep of DelH-pSB6A1(colony 3)
| Template | 1 µl of 1:1000 diluted M3 (pink) | 1 µl of 1:1000 diluted M4 (slightly pink) | 1 µl of 1:1000 diluted M5 (slightly pink) | 1 µl of 1:1000 diluted M6 (pink) | 1 µl of 1:1000 diluted M7 (pink) | 1 µl of 1:1000 diluted M 8 | 1 µl of 1:1000 diluted M 9 | 1 µl of 1:1000 diluted M10 | 1 µl of 1:1000 diluted M11 | 1 µl of 1:1000 diluted M12 |
|---|---|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature[°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Colonies 3 to 12 show expected size.
- => Perform miniprep and test digest with colonies 3-12.
Test Restriction Digest
Of colonies 3, 4, 5, 6, 7
| Component | Amount [µl] |
|---|---|
| DNA of M 3 to 12 (1:10) | 19 |
| CutSmart Buffer (10x) | .,2 |
| Enzyme (SalI) | 1 |
| ddH2O | - |
| Expected bands | 14,180 & 9,469 bp |
- Incubated over night at 37°C and shaking
Result
Expected bands: 14.18 and 9.459 Kb

l1:1 Kb+ ladder, l2:Digested miniprep of colony 3, l3:Digested miniprep of colony 4, l4:Digested miniprep of colony 5, l5:Digested miniprep of colony 6, l6:Digested miniprep of colony 7
l2-6:show not the expected fragment, but a band at ~ 4.8 Kb. Also observed pale bands at 20 Kb and ~ 3.8 Kb
None of the colonies shows expected restriction fragments. Additionally, there are pale bands at +20 Kb and ~3.8 Kb, but not the expected bands.
- => Possible explanation: colonies harbor reconjugated backbone AND something else involving DelH.
PCR Conditions CP.W17.B
M3 = Miniprep of DelH-pSB6A1(colony 3)
| Template | 1 µl of 1:1000 diluted M3 (pink) | 1 µl of 1:1000 diluted M4 (slightly pink) | 1 µl of 1:1000 diluted M5 (slightly pink) | 1 µl of 1:1000 diluted M6 (pink) | 1 µl of 1:1000 diluted M7 (pink) | 1 µl of 1:1000 diluted M 8 | 1 µl of 1:1000 diluted M 9 | 1 µl of 1:1000 diluted M10 | 1 µl of 1:1000 diluted M11 | 1 µl of 1:1000 diluted M12 |
|---|---|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Gel shows unexpected fragments at ~3 Kb.
- => Clones are negative despite of latest results. Current strategy does not work out. Development new strategy.
New Gibson Strategy
Gibson Strategy without mRFP
Try a new construct without mRFP, so that we can exclude the red clones from the screening. Therefore we are going to use a new reverse primer for the Backbone which includes only the terminator of the mRFP, but not the mRFP. The primers for the Backbone are HM11 & HM17. The construct is named pHM04 and shown in week 16 were the idea was specified.
For first approach pHM04, we will amplify the backbone without mRFP, avoiding backbone reassembly due to ribosome binding site homology. In the second strategy pHM05, we will additionally introduce a tetracycline resistance, to ensure integration of the insert.
Vector Maps and Primers
1st strategy: DelH & pSB6A1 without mRFP
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| HM17:DelH_Terminator_BB_fw | 16-08-2013 | Amplification of Backbone pSb6A1-lacI-mRFP | ATTGGCGCTGGAGTACGCGCTGGACTGA aggcatcaaataaaacgaaaggctcag |
2nd strategy: DelH & tetracycline resistance & pSB6A1 without mRFP
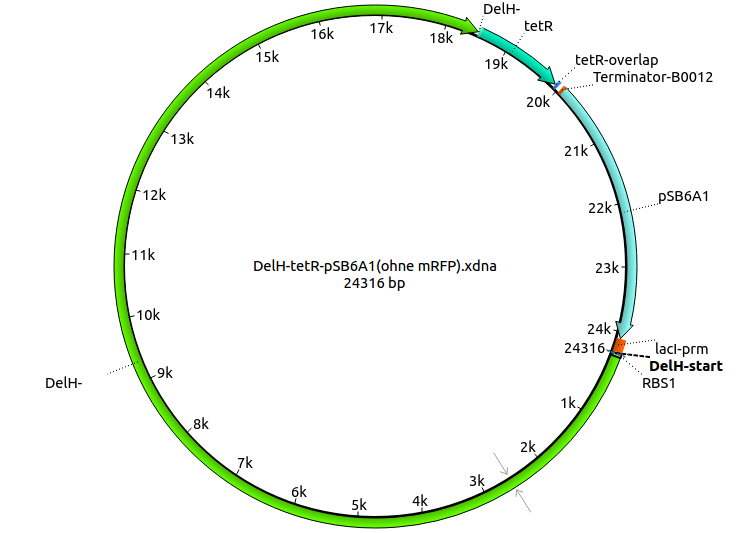
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| HM14:DelH_tetR_fw | 2013-08-16 | Gibson-Primer DelH-tetR: amplifies the tetracycline resistance from the pSB1T3 Backbone and creates an overlap to the end of DelH | ATTGGCGCTGGAGTACGCGCTGGACTGA atgaagttttaaatcaatctaaag |
| HM15:tetR_stop_BB_rev | 2013-08-16 | Gibson-Primer tetR-pSB6A1: amplifies the tetracycline resistance and creates an overlap with the Terminator of the Backbone pSB6A1 | Cgactgagcctttcgttttatttgatgcctggc ctcgtgatacgcctatttttatagg |
| HM16:tetR_pSB6A1_fw | 2013-08-16 | Gibson-Primer DelH, amplifies the Backbone pSB6A1 creating an overlap with the tetracycline resistance | Aaaaataggcgtatcacgag gccaggcatcaaataaaacgaaaggctcag |
Amplification of Tetracycline Fragment
Preparation of Tetraycline Medium
- Weight 100 mg of tetracycline and dissolve it in 10 ml ethanol = stock solution
- Add 500 µl of stock solution in 100 ml LB and mix
Transformation of Tetrazyklin Backbone
- For the new strategy we need a tetracycline resistance (length ~1.2 Kb) of the pSB1T3 Backbone (2013 Distribution, Plate 5, well 7A, insert: BBa_J04450) [http://parts.igem.org/Part:pSB1T3:Design|pSB1T3]
- The primers are already designed and shown in the table above
- With these primers, we amplify the tetracycline resistance and include an overlap to DelH end and the backbone pSB6A1 excluding the mRFP
- For the transformation I incubated 10 min at room temperature the well 7A of plate 5 with 10 µl ddH2O
PCR Conditions TR.W17.A
| Reagent | pSB1T3 | pSB1T3 |
|---|---|---|
| Template | picked colony 1 | picked colony 2 |
| Primer fw 10 µM | 1 µl HM14 | 1 µl HM14 |
| Primer rev 10 µM | 1 µl HM15 | 1 µl HM15 |
| Phusion Ready Mix | 10 µl | 10 µl |
| ddH2O | 8 µl | 8 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0,5°C) | 5 | |
| 72 | 30 | |
| 18 | 98 | 1 |
| 67 | 5 | |
| 72 | 30 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 1.468 Kb
Gel does not schow any band.
- => Try amplification with lower annealing temperature.
PCR Conditions TR.W17.B
| Reagent | pSB1T3 |
|---|---|
| Template | picked colony |
| Primer fw 10 µM | 1 µl HM14 |
| Primer rev 10 µM | 1 µl HM15 |
| Phusion Ready Mix | 10 µl |
| ddH2O | 8 µl |
| DMSO | - |
| Cycles | Temperature DelH BB [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 55 (touchdown -0,5°C) | 5 | |
| 72 | 30 | |
| 18 | 98 | 1 |
| 55 | 5 | |
| 72 | 30 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 1.468 Kb
Gel does not show expected band.
- => Repeat amplification with miniprep, therefore miniprep the 3 colonies containing pSB1T3.
PCR Conditions TR.W17.C
| Reagent | pSB1T3 | pSB1T3 | pSB1T3 |
|---|---|---|---|
| Template | Minipreped pSB1T3 | Minipreped pSB1T3 | Minipreped pSB1T3 |
| Primer fw 10 µM | 2 µl HM14 | 2 µl HM14 | 2 µl HM14 |
| Primer rev 10 µM | 2 µl HM15 | 2 µl HM15 | 2 µl HM15 |
| Phusion Ready Mix | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 55 (touchdown -0,5°C) | 5 | |
| 72 | 30 | |
| 18 | 98 | 1 |
| 55 | 5 | |
| 72 | 30 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 1.468 Kb

l1: tetracycline resistance amplified from minipreped colony 1, l2: tetracycline resistance amplified from minipreped colony 2, l3: tetracycline resistance amplified from minipreped colony 3,l4: 2 log,
l1-3: show expected band = was cut
Gel does show expected band.
- => Fragment was cut and gel extracted (c= 132.1 ng/µl).
Amplification of Backbone
PCR Conditions BB.W7.A
With new primers (HM16 & HM17)
| Reagent | BB with tetracycline & without mRFP | BB without mRFP |
|---|---|---|
| Template | 1 µl BB 3.08 | 1 µl BB 3.08 |
| Primer fw 10 µM | 2 µl HM11 | 2 µl HM11 |
| Primer rev 10 µm | 2 µl HM16 | 2 µl HM17 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O 5µl | 5 µl |
| Cycles | Temperature DelH BB [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 69 (touchdown -0,5°C) | 5 | |
| 72 | 75 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 75 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: ~ 4.4 Kb
Gel shows specific band for amplification using HM16.
- => Fragment wa scut and gel extracted. For HM17, run a two-step PCR because of it's high annealing temperature. Also we are going to rise the amplification time to 1:30 min.
PCR Conditions BB.W7.B
| Reagent | BB with tetracycline & without mRFP | BB without mRFP | BB with tetracycline & without mRFP | BB without mRFP |
|---|---|---|---|---|
| Template | 1 µl BB 3,08 | 1 µl BB 3,08 | 1 µl BB 3,08 | 1 µl BB 3,08 |
| Primer fw 10 µM | 2 µl HM11 | 2 µl HM11 | 5 µl HM11 | 5 µl HM11 |
| Primer rev 10 µM | 2 µl HM16 | 2 µl HM17 | 5 µl HM16 | 5 µl HM17 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 25 µl | 25 µl |
| ddH2O | 5 µl | 5 µl | 14 µl | 14 µl |
| Cycles | Temperature (20 µl = sample 1 & 2) [°C] | Time | Temperature (50 µl = sample 3 & 4) [°C] | Time [s] |
|---|---|---|---|---|
| 1 | 98 | 5 | 98 | 5 |
| 12 | 98 | 1 | 98 | 1 |
| 69 (touchdown -0,5°C) | 5 | - | ||
| 72 | 90 | 72 | 90 | |
| 18 | 98 | 1 | 98 | 1 |
| 68 | 5 | - | ||
| 72 | 90 | 72 | 90 | |
| 1 | 72 | 5 min | 72 | 5 min |
| 1 | 4 | inf | 4 | inf |
Result
Expected band: ~ 4.4 Kb
All samples show expected length.
- => Fragments were cut and gel extracted.
| Sample | Concentration [ng/µl] |
|---|---|
| BB-16 | 76.7 |
| BB-17 | 51.3 |
Restriction Digest with DpnI
- Incubated at 37°C for 2.5 h
| Sample [µl] | Buffer [µl] | Enzyme [µl] |
|---|---|---|
| 18 BB-16 | 2.1 CutSmart Buffer | 1 DpnI |
| 18 BB-16 | 2.1 CutSmart Buffer | 1 DpnI |
- Afterwards, a purification was performed with the nucleotide removal kit
Generation of DelH Plasmid pHM04 23-08
Summary of Fragments
| Fragment | Concentration [ng/µl] | Date |
|---|---|---|
| G0 | 6.4 | 22-08 |
| G1/2a | 8.5 | 22-08 |
| G2b | 18.6 | Example |
| BB | Example | 22-08 |
Gibson Assembly
| Mix name | DelH G0 | DelH G1/2a | DelH G2b | BB17 | Gibson Master Mix [µl] | Final volume [µl] |
|---|---|---|---|---|---|---|
| E1 | 9 | - | - | 0.5 | 10 | 20 |
| E2 | - | 7 | 2.5 | 0.5 | 10 | 20 |
- Incubate the Gibson Assembly for 1 h at 50°C
Electroporation
| Sample | Electroporated with | Plated out | Cuvette "exploded" |
|---|---|---|---|
| 1 | 1 µl of E1 (5 µl Gibson + 10 µl ddH2O) | 22.08.2013 | v |
| 2 | 14 µl of E1 (5 µl Gibson + 10 µl ddH2O) | 22.08.2013 | v |
| 3 | 1 µl of E2 (5 µl Gibson + 10 µl ddH2O) | 22.08.2013 | - |
| 4 | 14 µl of E2 (5 µl Gibson + 10 µl ddH2O) | 22.08.2013 | v |
| 5 | 20 µl of E1 (purified by isoprop-precipitation) | 22.08.2013 | - |
| 6 | 20 µl of E2 (purified by isoprop-precipitation) | 22.08.2013 | - (no pellet observed) |
Colony-PCR CP.W17.C
- 50 colonies of plate 5 (= isoprop purified E1 => G0-complete in Backbone)
- 10 colonies of plate ...
- 2 different screening are performed
- PC = picked colony
- S5 = Sample 5 (watch names above on construct DelH-BB)
| Template | 50 x 1 PC S5 | 50 x 1 PC S5 | 4 x 1 PC S3 | 4 x 1 PC S3 |
|---|---|---|---|---|
| Expected length [bp] | 663 | 2,500 | 663 | 2,500 |
| Named | A -J (5 colonies per tube) | A -J (5 colonies per tube) | 1-4 | 1-4 |
| Primer fw 10 µM | 10 x 1 µl VF2 | 10 x 1 µl HM13 | 4 x 1 µl VF2 | 4 x 1 µl HM13 |
| Primer rev 10 µM | 10 x 1 µl DN07 | 10 x 1 µl VR2 | 1 µl DN07 | 1 µl VR2 |
| Dream-Taq Polymerase (2x) | 10 x 10 µl | 10 x 10 µl | 4 x 10 µl | 4 x 10 µl |
| ddH2O | 10 x 8 µl | 10 x 8 µl | 4 x 8 µl | 4 x 8 µl |
| Cycles | Temperature Screening start[°C] | Time [s] | Temperature Screening end [°C] | Time [s] |
|---|---|---|---|---|
| 1 | 95 | 120 | 95 | 120 |
| 12 | 95 | 60 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | 60 (touchdown -0.5°C) | 30 | |
| 72 | 45 | 72 | 2:30 min | |
| 12x/ 34x | 95 | 60 | 95 | 60 |
| 65 | 30 | 60 | 30 | |
| 72 | 45 | 72 | 2:30 min | |
| 1 | 72 | 5 min | 72 | 5 min |
| 1 | 12 | inf | 12 | inf |
Result
Expected band: 663 bp (screening start), 2.5 Kb (screening end)
Gel does not show expected bands.
- => Screen 10 minipreped colonies of the plate 5 and pick another 10 colonies of plate 1, 2, 3 and 6 (on plate 4 are no colonies).
Colony-PCR CP.W17.D
- 2 different screening are performed
- PC = picked colony
- S5 = Sample 5 (watch names above on construct DelH-BB)
| Template | 10 x 1 PC S1 | 10 x 1 PC S1 | 10 x 1 PC S2 | 10 x 1 PC S2 | 10 x 1 PC S3 | 10 x 1 PC S3 | 10 x 1 PC S6 | 10 x 1 PC S6 | 10 x 1 µl of minipreped colony | 10 x 1 µl of minipreped colony |
|---|---|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 2.5 | 663 | 2.5 | 663 | 2.5 | 663 | 2.5 | 663 | 2.5 |
| Named | 1A-E (2 colonies per tube) | 1F-J (2 colonies per tube) | 2A-E (2 colonies per tube) | 2F-J (2 colonies per tube) | 3A-E (2 colonies per tube) | 3F-J (2 colonies per tube) | 6A-E (2 colonies per tube) | 6F-J (2 colonies per tube) | ............. | .................... |
| Primer fw 10 µM | 5 x 1 µl VF2 | 5 x 1 µl HM13 | 5 x 1 µl VF2 | 5 x 1 µl HM13 | 5 x 1 µl VF2 | 5 x 1 µl HM13 | 5 x 1 µl VF2 | 5 x 1 µl HM13 | 5 x 1 µl VF2 | 5 x 1 µl HM13 |
| Primer rev 10 µM | 5 x 1 µl DN07 | 5 x 1 µl VR2 | 5 µl DN07 | 5 µl VR2 | 5 x 1 µl DN07 | 5 x 1 µl VR2 | 5 µl DN07 | 5 µl VR2 | 5 x 1 µl DN07 | 5 x 1 µl VR2 |
| Dream-Taq Polymerase (2x) | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl |
| ddH2O | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 6 µl | 5 x 6 µl |
| Cycles | Temperature Screening start [°C] | Time [s] | Temperature Screening end [°C] | Time [s] |
|---|---|---|---|---|
| 1 | 95 | 120 | 95 | 120 |
| 12 | 95 | 60 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | 60 (touchdown -0.5°C) | 30 | |
| 72 | 45 | 72 | 2:30 min | |
| 12x/ 34x | 95 | 60 | 95 | 60 |
| 65 | 30 | 60 | 30 | |
| 72 | 45 | 72 | 2:30 min | |
| 1 | 72 | 5 min | 72 | 5 min |
| 1 | 12 | inf | 12 | inf |
Result
Expected band: 663 bp (screening start), 2.5 Kb (screening end)
None of the gels shows expected fragment.
- => Overthink strategy.
Amplification of 1
26-08 - 01-09-13
Generation of DelH Plasmid pHM05 26-08
Overview on Fragments
| Fragment | Concentration [ng/µl] | Date |
|---|---|---|
| G0 | 6.4 | 22-8 |
| G1/2a | 8.5 | 22-08 |
| G2b | 18.6 | 09-08 |
| BB | 40.6 | 22-08 |
| TetR | 132.1 | 26-08 |
Gibson Assembly
| Mix name | DelH-G0 | DelH-1/2a | DelH-G2b | BB17 | TetR (1:10) | GibsonMasterMix | Final volume |
|---|---|---|---|---|---|---|---|
| 0 | 8.5 µl | - | - | 0.5 µl | 1 µl | 10 µl | 20 µl |
| 1 | - | 6.5 µl | 2 µl | 0.5 µl | 1 µl | 10 µl | 20 µl |
- Incubate the Gibson Assembly for 1 h at 50°C in Thermocycler
Electroporation
In the next step, we purify 10 µl with Isoprop purification protocol and another 5 µl we dilute with ddH2O (10 µl)
| Sample | Amount | Used in Elektroporation |
|---|---|---|
| 0 A | 5 µl + 10 µl ddH2O |
|
| 0 B | 20 µl isoprop purified |
|
| 1 A | 5 µl + 10 µl ddH2O |
|
| 1 B | 20 µl isoprop purified |
|
Colony-PCR CP.W18.A
- 10 colonies of plate 1-8 (names are explained in the following table)
- PC = picked colony
- S5 = Sample 5 (watch names above on construct DelH-BB)
| Template | 10 x 1 PC S5 | 50 x 1 PC S5 | 4 x 1 PC S3 | 4 x 1 PC S3 |
|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 |
| Named | A -J (5 colonies per tube) | A -J (5 colonies per tube) | 1-4 | 1-4 |
| Primer fw 10 µM | 10 x 1 µl VF2 | 10 x 1 µl VF2 | 4 x 1 µl VF2 | 4 x 1 µl VF2 |
| Primer rev 10 µM | 10 x 1 µl DN07 | 10 x 1 1 µl DN07 | 1 µl DN07 | 1 1 µl DN07 |
| Dream-Taq Polymerase (2x) | 10 x 10 µl | 10 x 10 µl | 4 x 10 µl | 4 x 10 µl |
| ddH2O | 10 x 8 µl | 10 x 8 µl | 4 x 8 µl | 4 x 8 µl |
| Cycles | Temperature Screening start [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 14 | 95 | 60 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Colonies 15, 32, 33, 35, 39, 41, 42 and 46 show a shadow specific band at ~600 bp, slightly too low.
=> ?????
Amplification of DelH G0
Gradient-PCR Conditions G0.W18.A
| Reagent | G0 | G0 | G0 | G0 |
|---|---|---|---|---|
| Expected length [Kb] | 18 | 18 | 18 | 18 |
| Named | G0 65 | G0 66 | G0 67 | G0 68 |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 1 µl DN11 | 1 µl DN11 | 1 µl DN11 | 1 µl DN11 |
| Primer rev 10 µM | 1 µl HM08 | 1 µl HM08 | 1 µl HM08 | 1 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl |
| DMSO | 1 | 1 | 1 | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65/66/67/68 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb

l1: 1Kb+ ladder, l2: G0 amplified at 65°C, l3:G0 amplified at 66°C, l4:G0 amplified at 67°C, l5: G0 amplified at 68°C,
l2-5: show slightly band at expected 18 Kb, the 5 Kb band is observed at every temperature, the best temperature seems to be 67°C
Gel shows unspecific band at 5 Kb. G0 was best amplified at 67°C.
- => Fragment was cut and gel isolated.
Re-PCR Conditions G0.W18.B
| Reagent | G0 | G0 |
|---|---|---|
| Expected length [Kb] | 18 | 18 |
| Named | G0 | G0 1 |
| Template | 1 µl gel extracted G0 from 22-08 | 1 µl gel extracted G0 from 26-08 |
| Primer fw 10 µM | 1 µl DN11 | 1 µl DN11 |
| Primer rev 10 µM | 1 µl HM08 | 1 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl |
| DMSO | 1 | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb
A slight band can be observed at 18 Kb, but 5Kb is much more amplified.
- => By NCBI-Blast we found out that primer HM08 can also bind inside of DelH with 6 mismatches and the forward primer DN11 can also bind with 4 mismatches inside the DelH = possible explanation for 5 Kb.
- => Change the amplification conditions.
PCR Conditions G0.W18.C
| Reagent | G0 | G0 | G0 |
|---|---|---|---|
| Expected length [Kb] | 18 | 18 | 18 |
| Named | 0 | 1 | 2 |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 1 µl DN11 | 1 µl DN11 | 1 µl DN11 |
| Primer rev 10 µM | 1 µl HM08 | 1 µl HM08 | 1 µl HM08 |
| Phusion II Polymerase (Hot Start) | 0.2 µl | 0.2 µl | 0.2 µl |
| dNTPs | 0.4 µl | 0.4 µl | 0.4 µl |
| GC buffer (5x) | 4 µl | 4 µl | 4 µl |
| ddH2O | 12.4 µl | 11.4 µl | 10.4 µl |
| DMSO | 0 µl | 1 µl | 2 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 10 |
| 65 | 20 | |
| 72 | 5:00 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Result
Expected band: 18 Kb
Only PCR with 2 µl DMSO shows any band, but not the expected at 18 Kb, only the band at 5 Kb.
- => Further improve PCR condtions.
Amplification of DelH G1/2a
Re-PCR Conditions G1/2a.W18.A
| Reagent | G1/2a |
|---|---|
| Expected length [Kb] | 18 |
| Named | G1/2a |
| Template | 1 µl gel extracted G0 from 22-08 |
| Primer fw 10 µM | 1 µl DN11 |
| Primer rev 10 µM | 1 µl HM06 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 6 µl |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb
Expected fragment was amplified, but gel shows numerous side bands.
- => Fragment was cut and gel isolated.
Gradient-PCR Conditions G1/2a.W18.A
| Reagent | G1/2a | G1/2a | G1/2a | G1/2a |
|---|---|---|---|---|
| Expected length [Kb] | 13 | 13 | 13 | 13 |
| Named | 1 | 2 | 3 | F |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl gel extracted G1/2a fragment |
| Primer fw 10 µM | 1 µl DN11 | 1 µl DN11 | 1 µl DN11 | 1 µl DN11 |
| Primer rev 10 µM | 1 µl HM06 | 1 µl HM06 | 1 µl HM06 | 1 µl HM06 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 5 µl | 4 µl | 6 µl |
| DMSO | 1 | 2 | 3 | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 67 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
- => There is no band at 18 Kb visible - entire DelH fragment could not be amplified.
Amplification of DelH FS64 - FS71
PCR Conditions FS64-FS71.W18.A
| Reagent | DN11-FS66 | FS67-FS68 | FS69-HM08 | FS69-FS70 | FS71-HM08 | FS67 -HM08 |
|---|---|---|---|---|---|---|
| Expected length [Kb] | 4.7 | 4.0 | 9.7 | 7.0 | 2.8 | 14.0 |
| Named | 1 | 2 | 3 | 4 | 5 | 6 |
| Template | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl DN11 | 2.5 µl FS67 | 2.5 µl FS69 | 2.5 µl FS69 | 2.5 µl FS71 | 2.5 µl FS67 |
| Primer rev 10 µM | 2.5 µl FS66 | 2.5 µl FS68 | 2.5 µl HM08 | 2.5 µl FS70 | 2.5 µl HM08 | 2.5 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 3 µl | 3 µl | 3 µl | 3 µl | 3 µl | 3 µl |
| DMSO | 1 | 1 | 1 | 1 | 1 | 1 |
| Cycles | Temperature DN11-FS66 [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68↓ | 5 | |
| 72 | 1:25 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 85 | |
| 1 | 72 | 6 min |
| 1 | 10 | inf |
| Cycles | Temperature [°C] | Time FS67 - FS68 | Time FS69-HM08 | Time FS69-FS70 | FS67 -HM08 | Time FS71-HM08 |
|---|---|---|---|---|---|---|
| 1 | 98 | 5 | 5 | 5 | 5 | 5 |
| 12 | 98 | 1 | 1 | 1 | 1 | 1 |
| 66↓ | 5 | 5 | 5 | 5 | 5 | |
| 72 | 70 | 3:00 min | 2:10 min | 4:38 min | 52 | |
| 18 | 98 | 1 | 1 | 1 | 1 | 1 |
| 64 | 5 | 5 | 5 | 5 | 5 | |
| 72 | 70 | 3:00 min | 2:10 min | 4:38 min | 52 | |
| 1 | 72 | 5 min | 10 min | 5 min | 5 min | 5 min |
| 1 | 10 | inf | inf | inf | inf | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected bands: DN11-FS66 = 4.7 Kb, FS67-FS68 = 4.0 Kb, FS69-HM08 = 9.7 Kb, FS69-FS70 = 7.0 Kb, FS71-HM08 = 2.7 Kb, FS67-HM08 = 14 Kb
Gel shows expected fragments of DN11-FS66, FS67-FS68 and FS71-HM08
- => Bands were cut. Run gel with remaining sample for gel extraction.
- => Optimize remaining PCRs.
Amplification of DelH DN11-FS66
PCR Conditions DN11-FS66.W18.A
| Reagent | 4x DN11 - FS66 |
|---|---|
| Expected length [Kb] | 4.7 |
| Named | 1 |
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl DN11 |
| Primer rev 10 µM | 2.5 µl FS66 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 3 µl |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68↓ | 5 | |
| 72 | 85 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 85 | |
| 1 | 72 | 6 min |
| 1 | 10 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 4.7 Kb
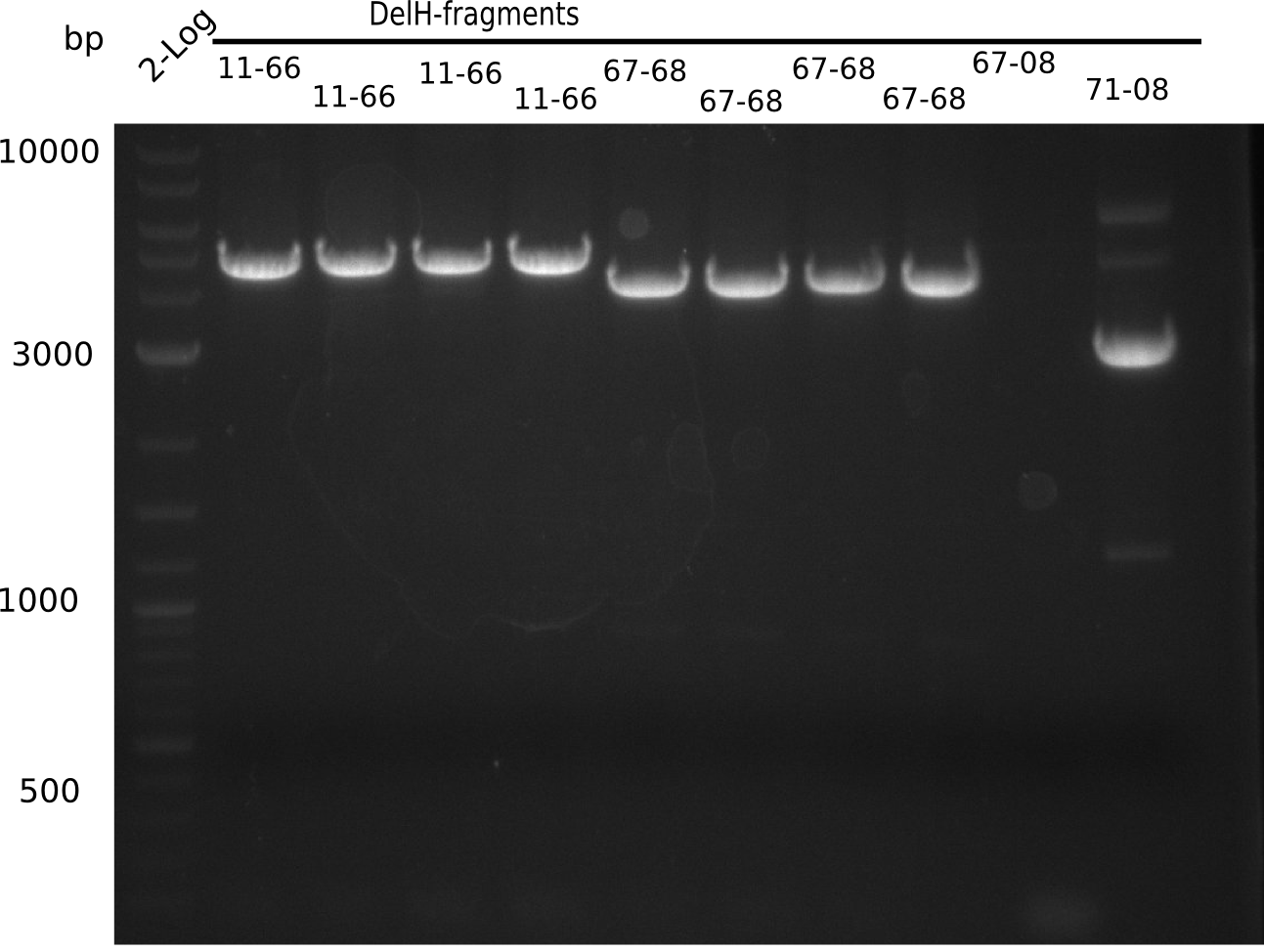
l1:2log ladder, l2-5: DelH amplified with DN11 & FS66, l6-9:DelH amplified with FS67 & FS68, l10:DelH amplified with FS67 & HM08, l11:DelH amplified with FS71 & HM08
l2-5: shows expected band at 4.7 Kb, l6-9: at 4.0 Kb, l10 doesn't show band at 14 Kb, l11:show band at 2.7 => all specific bands were cut out
Bands could not be used, because too much UV light was applied.
Amplification of DelH FS67-FS68
PCR Conditions FS67-FS68.W18.A
| Reagent | 4x FS67-FS68 |
|---|---|
| Expected length [Kb] | 4.0 |
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl FS67 |
| Primer rev 10 µm | 2.5 µl FS68 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 3 µl |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66↓ | 5 | |
| 72 | 1:10 min | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 70 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 4.0 Kb

l1:2log ladder, l2-5: DelH amplified with DN11 & FS66, l6-9:DelH amplified with FS67 & FS68, l10:DelH amplified with FS67 & HM08, l11:DelH amplified with FS71 & HM08
l2-5: shows expected band at 4.7 Kb, l6-9: at 4.0 Kb, l10 doesn't show band at 14 Kb, l11:show band at 2.7 => all specific bands were cut out
Bands could not be used, because too much UV light was applied.
Amplification of DelH FS71-HM08
PCR Conditions FS71-HM08.W18.A
| Reagent | FS71-HM08 |
|---|---|
| Expected length [Kb] | 2.8 |
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl FS71 |
| Primer rev 10 µM | 2.5 µl HM08 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 3 µl |
| DMSO | 1 |
| Cycles | Temperature A [°C] | Time | Temperature B [°C] | Temperature C [°C] |
|---|---|---|---|---|
| 1 | 98 | 5 | 98 | 98 |
| 12 | 98 | 1 | 98 | 98 |
| 68↓ | 5 | - | 69 | |
| 72 | 52 | 72 | 72 | |
| 18 | 98 | 1 | 98 | 98 |
| 66 | 5 | - | 69 | |
| 72 | 52 | 72 | 72 | |
| 1 | 72 | 6 min | 72 | 72 |
| 1 | 10 | inf | 10 | 10 |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 2.8 Kb

l1:2log ladder, l2-5: DelH amplified with DN11 & FS66, l6-9:DelH amplified with FS67 & FS68, l10:DelH amplified with FS67 & HM08, l11:DelH amplified with FS71 & HM08
l2-5: shows expected band at 4.7 Kb, l6-9: at 4.0 Kb, l10 doesn't show band at 14 Kb, l11:show band at 2.7 => all specific bands were cut out
Bands could not be used, because too much UV light was applied.
PCR Conditions FS71-HM08.W18.B
| Reagent | FS71-HM08 |
|---|---|
| Expected length [Kb] | 2.8 |
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl FS71 |
| Primer rev 10 µM | 2.5 µl HM08 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 3 µl |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 52 | |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 52 | |
| 1 | 72 | 6 min |
| 1 | 10 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 2.8 Kb
Both condistions resulted in the expected fragment.
- => Fragments were cut and gel extracted.
- => Run new PCR with 72.3°C constant annealing temperature because of the slight side bands.
Amplification of DelH FS69-FS70
PCR Conditions FS69-FS70.W18.A
| Reagent | FS69-FS70 |
|---|---|
| Expected length [Kb] | 7.0 |
| Named | FS69-FS70 |
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl FS69 |
| Primer rev 10 µM | 2.5 µl FS70 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 3 µl |
| DMSO | 1 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66↓ | 5 | |
| 72 | 2:10 min | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 2:10 min | |
| 1 | 72 | 6 min |
| 1 | 10 | inf |
- Lid preheated at 98°C
- No hot start
Characterization of Amplified DelH Fragments
Test Restriction Digest
| Fragment | Fragment length [Kb] | Digestes with following enzymes | Expected bands [Kb] |
|---|---|---|---|
| DN11-FS66 | 4.7 | EcoRV | 3.7 & 0.9 |
| FS67-FS68 | 4.0 | NotI | 2.7 & 1.3 |
| FS69-FS70 | 7.0 | SalI | 5.5 & 1.6 |
| FS71-HM08 | 2.8 | NotI & HindIII | 1.3 & 1.1 & 0.3 |
| Fragment | Amount in digest | Enzyme | Buffer | ddH2O | Final amount |
|---|---|---|---|---|---|
| DN11-FS66 | 5 µl | 1 µl EcoRV | 2 µl 3.1 buffer | 10.9 µl | 20 µl |
| FS67-FS68 | 5 µl | 1 µl NotI | 2 µl 3.1 buffer | 10.9 µl | 20 µl |
| FS69-FS70 | 5 µl | 1 µl SalI-HF | 2 µl CutSmart buffer | 12 µl | 20 µl |
| FS71-HM08 | 5 µl | 1 µl NotI-HF & HindIII-HF | 2 µl CutSmart buffer | 11 µl | 20 µl |
Result
Expected banda are listed above.
The test digest showed that the first fragment is cut, because the band is around 3.7 Kb, but the third and fourth lane on the gel picture show the complete non-digested fragment and on line 5 the FS71-HM08 fragment is not visible.
- => check again the digests.
Generation of DelH Plasmid pHM04 and pHM05 30-08
Gibson Assembly
| Fragment | Concentration [ng/µl] | Amount with BB16 & tetR [µl] | Amount with BB17 [µl] | Amount with mRFP BB of pHM03 |
|---|---|---|---|---|
| DN11-FS66 | 172.5 | 1.67 | 1.47 | 1.67 |
| FS67-FS68 | 120.7 | 2.03 | 1.79 | 2.03 |
| FS69-FS70 | 211.3 | 2.03 | 1.79 | 2.03 |
| FS71-HM08 | 146 | 1.18 | 1.04 | 1.18 |
| BB16 | 40.3 | 2.54 | 0 | 0 |
| tetR (1:10) | 13.2 | 0.56 | 0 | 0 |
| BB17 | 23.0 | 0 | 3.91 | 0 |
| BB+mRFP (pHM03) | 0 | 0 | 0 | 3.08 |
Electroporation of pHM04
10 µl H2O were added to 5 µl of the Gibson assembly mix. 1 µl and 14 µl were electroporated into electrocompetent DH10ß. Cells were incubated at 37°C for 1 h and streaked (10% vs. 90%) on LB Amp plates and stored ON at 37°C.
Colony-PCR Conditions CP.W18.B
14 colonies (#7 was left out) were picked and screened for correct integration of first DelH fragmenta s well as grown in 500 µl LB Amp.
| Template | DelH construct pHM04 30-08 |
|---|---|
| Expected length [bp] | 663 |
| Named | Colonies 1 - 15 |
| Primer 10 µM fw | 2 µl VF2 |
| Primer 10 µM rev | 2 µl DN07 |
| Dream-Taq Polymerase (2x) | 10 µl |
| ddH2O | 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 66 (touchdown -0.5°C) | 30 | |
| 72 | 2:30 min | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 2:30 min | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Colonies 1, 3, 4, 6, 12 and 15 show positive band.
- =>Of the 250 µl liquid culture, screening PCRs for the correct insertion of the last DelH fragment were performed. To the remaining 250 µl, 8 ml LB Amp were added. Colonies 1 and 3 did not grow and were excluded from further analysis.
Colony-PCR CP.W18.C
| Template | pHM04 30-08 |
|---|---|
| Expected length [Kb] | 2.5 |
| Named | Colonies 1 - 15 |
| Primer 10 µM fw | 2 µl HM13 |
| Primer 10 µM rev | 2 µl VR |
| Dream-Taq Polymerase (2x) | 10 µl |
| ddH2O | 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 2:30 min | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 2:30 min | |
| 1 | 12 | inf |
Results
Expected band: 2.5 Kb
Colonies 4, 6, 12 and 15 show positive band.
=> Of 2 ml of the cultures, glycerol stocks were prepared. 3 ml were mini preped and test digested. The remaining 3 ml were stored.
Test Restriction Digest of Colony S4
| Clone | Concentration [ng/µl] | Amount in digest | Enzyme | Buffer | ddH2O | Final amount |
|---|---|---|---|---|---|---|
| pHM04 S4 | 400 ng/µL | 1 µl | 1 µl EcoRV | 1 µl CutSmart buffer | 8 µl | 10 µl |
| pHM04 S4 | 400 ng/µL | 1 µl | 1 µl NotI | 1 µl CutSmart buffer | 8 µl | 10 µl |
| pHM04 S4 | 400 ng/µL | 1 µl | 1 µl SalI-HF | 1 µl CutSmart buffer | 8 µl | 10 µl |
Result
Expected bands are shown in table below.
| Clone | Clone length [bp] | Digestes with following enzymes | Expected bands [bp] |
|---|---|---|---|
| pHM04 S4 | 22,937 | EcoRI | 17,829 & 5,108 |
| pHM04 S4 | 22,937 | NotI | 11,079, 6201, 3,998 & 1,628 |
| pHM04 S4 | 22,937 | PvuI | 11,621, 8628 & 2,685 |
- The expected bands were not there. So the colony seems not to have the entire DelH-plasmid. .
Amplification of 1
02-09 - 08-09-13
Characterization of DelH Plasmid pHM04 30-08 Clone 12
Maxiprep
- 1 ml of an ON culture DH10ß-DelH were inocculated in 200 ml LB Amp and cultured ON at 37°C.
- Maxi prep was performed by upscaling of mini precipitation protocol.
Result
The maxi prep yielded 6x 180 µl.
| Maxi prep | Concentration [µg/µl] |
|---|---|
| 1 | 5.40 |
| 2 | 5.37 |
| 3 | 5.42 |
| 4 | 5.40 |
| 5 | 5.38 |
| 6 | 5.42 |
- Sequencing showed a mutation at the beginning of DelH
- Next steps
- => Sequencing the other 3 positive colonies => hopefully one of them has no mutation at the beginning of DelH
- => Sequence D. acidovorans, maybe the organism SPH1 was not perfectly sequenced => amplify with FS35 & DN07
Characterization of DelH Plasmid pHM04 30-08 Clones 4, 6, 15
Sequencing
- Miniprep performed
- Send in for sequencing => no reliable result
- Maxiprep performed => Clone 4: c= 5.51 µg/µl, Clone 6: c= 3.71µg/µl, Clone 15: c= 4.65 µg/µl
- Send in for sequencing with DN07 again
Result
| Sample | Alignment File |
|---|---|
| Midi-col 4 | File:Heidelberg Sequencing Result pHM04 DN07 colony 04.clustal.txt |
| Midi-col 6 | sequencing insufficient |
| Maxi-col 15 | sequencing insufficient |
Electroporation
Because the sequenced DelH colony 12 has a mutation at the beginning of the coding DNA, we are sequencing the other colonies and parallel we transformed with the rest of the same Gibson Mix (15 µl) used for the electroporation before (colonies 4, 6, 12, 15). In advance, we purified it via isoprop purification protocol.
| Sample | Isoprop purified Gibson Mix | Colonies grown? |
|---|---|---|
| A | 10 µl | yes |
| B | 20 µl | yes (a lot more than on sample A) |
Colony-PCR conditions CP.W19.A
There were 30 colonies screened. 3 per tube and in total 10 colonies of sample A & 20 colonies of sample B
| Template | 3 PC of S-A | 3 PC of S-A | 4 PC of S-A | 3 PC of S-B | 3 PC of S-B | 4 PC of S-B | 3 PC of S-B | 3 PC of S-B | 4 PC of S-B |
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 1-3 | 4-6 | 7-10 | 11-13 | 14-16 | 17-20 | 21-23 | 24-26 | 27-30 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 |
| iTaq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
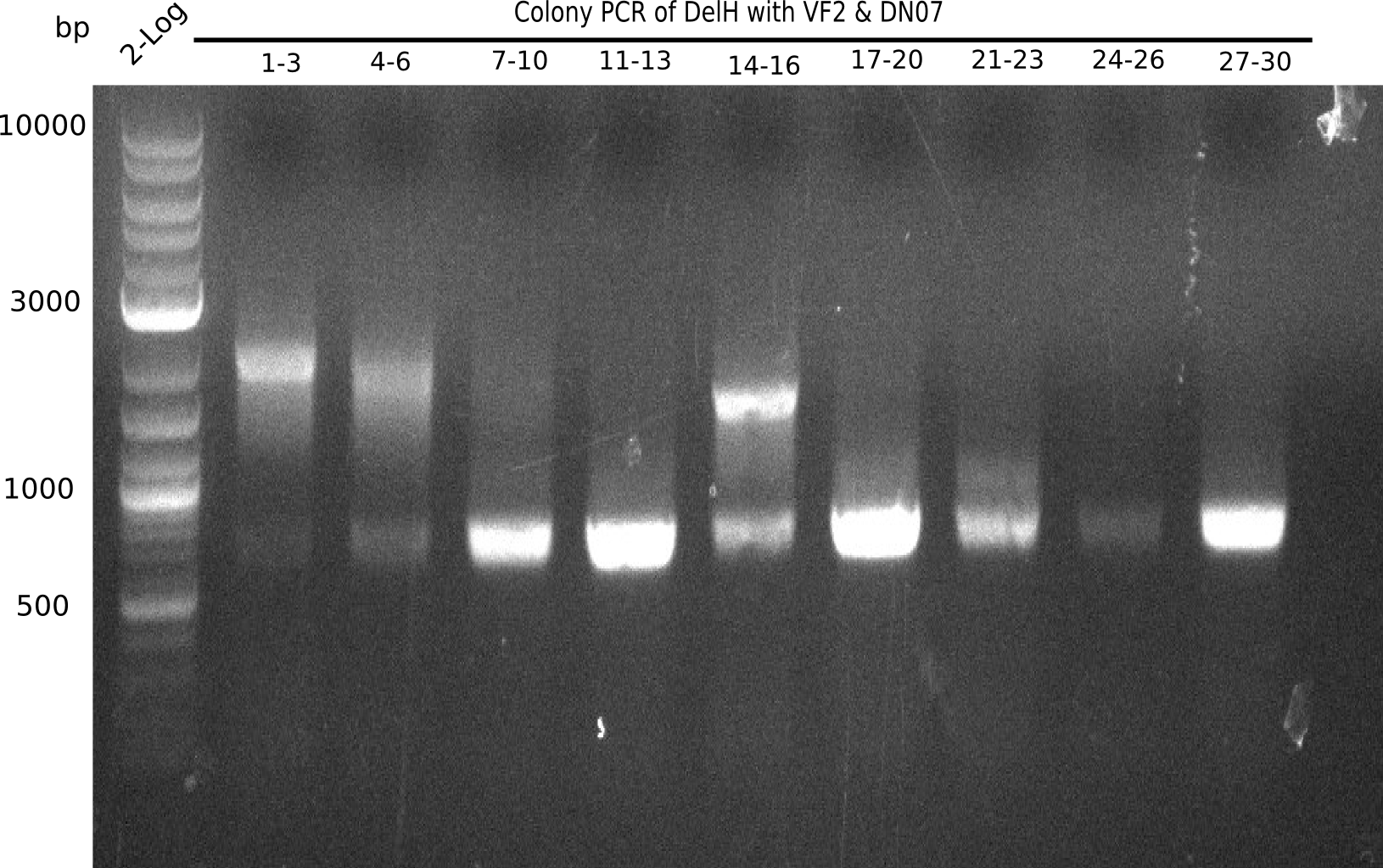
l1:2log, l2:colonies 1-3, l3:colonies 4-6, l4:colonies 7-10, l5:colonies 11-13, l6:colonies 14-16, l7:colonies 17-20, l8:colonies 21-23, l9:colonies 24-26, l10:colonies 27-30
l4, l5,l8,l10:show the specific band at 663 bp
Some of the colony mixes show positive band.
- => Screen again colonies 7-10, 11-13, 17-20, 27-30 in a single PCR tubes.
Colony-PCR conditions CP.W19.B
| Template | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 17 | 18 | 19 | 20 | 27 | 28 | 29 | 30 |
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 17 | 18 | 19 | 20 | 27 | 28 | 29 | 30 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 |
| iTaq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Colonies 7,13, 19 & 29 show a definit result
- => Add medium and perform a mediprep the next day for sending in for sequencing.
Midiprep
Performed with 300 µl P1, 600 µl P2 and after incubating for 180 450 µl S3. Protocol was performed with isoprop/ethanol purification.
| Sample | Concentration [ng/µl] |
|---|---|
| Midi-col 7 | 2628.5 |
| Midi-col 13 | 2379.3 |
| Midi-col 19 | 2846.6 |
| Midi-col 29 | 2846.6 |
Test Restriction Digest
We performed a test digest with EcoRI-HF of the screened samples col 7, col 13, col 19, col 29. We expect a band of 17.9 Kb and 4.9 Kb.
| Sample | Concentration [ng/µl] | Amount in restriction digest | EcoRI-HF [µl] | CutSmartBuffer [µl] | ddH2O [µl] | Total amount [µl] |
|---|---|---|---|---|---|---|
| Midi-col 7 | 2628.5 | 0.5 µl = 1314.25 ng | 1 | 2 | 16.5 | 20 |
| Midi-col 13 | 2379.3 | 0.5 µl = 1189.65 ng | 1 | 2 | 16.5 | 20 |
| Midi-col 19 | 2846.6 | 0.5 µl = 1423.3 ng | 1 | 2 | 16.5 | 20 |
| Midi-col 29 | 2846.6 | 0.5 µl = 1423.3 ng | 1 | 2 | 16.5 | 20 |
- Incubation time: 1 h at 37 °C
Result
Expected bands: 17.9 Kb and 4.9 Kb
Colonies 7 & 13 show a definit result
- => Add medium and perform a mediprep the next day for send in for sequencing.
Sequencing
The colonies 7 & 19 were sent in MWG for sequencing. There for we prepared 15 µl of the plasmid (midiprep) with a concentration of 50-100 ng/µl and added 2 µl DN07 primer (10µM)
| Sample | Concentration [ng/µl] | Amount for sequencing | ddH2O added up to 15 µl | Final concentration [ng/µl] |
|---|---|---|---|---|
| Midi-col 7 | 2628.5 | 0.5 µl = 1314.25 ng | 14.5 | 87.62 |
| Midi-col 19 | 2846.6 | 0.5 µl = 1423.3 ng | 14.5 | 94.89 |
| Maxi-col 4 | 5500 | 2 µl of 1:10 dilution = 1100 ng | 13 | 73.3 |
Result
| Sample | Alignment File |
|---|---|
| Midi-col 7 | File:Heidelberg Sequencing Result pHM04 DN07 colony 7.clustal.txt |
| Midi-col 19 | File:Heidelberg Sequencing Result pHM04 DN07 colony 19-2.clustal.txt |
| Maxi-col 4 | File:Heidelberg Sequencing Result pHM04 DN07 colony 04.clustal.txt |
Test Restriction Digest
We performed a test digest with EcoRI-HF and PvuI-HF of the screened samples col 4, col 7, col 19. We expect a band of 17.9 Kb and 4.9 Kb for the digest with EcoRI-HF and bands of 11.5 Kb, 8.5 Kb and 2.6 Kb.
| Sample | Concentration [ng/µl] | Amount in restriction digest | Enzyme | CutSmartBuffer [µl] | ddH2O [µl] | Total amount [µl] |
|---|---|---|---|---|---|---|
| Midi-col 4 | 5500 | 0.25 µl = 1375 ng | EcoRI-HF 1 µl | 2 | 16.75 | 20 |
| Midi-col 4 | 5500 | 0.25 µl = 1375 ng | PvuI-HF 1 µl | 2 | 16.75 | 20 |
| Midi-col 7 | 2628.5 | 0.5 µl = 1314.25 ng | PvuI-HF 1 µl | 2 | 16.5 | 20 |
| Midi-col 19 | 2846.6 | 0.5 µl = 1423.3 ng | PvuI-HF 1 µl | 2 | 16.5 | 20 |
- Incubation time: 1:20 h at 37 °C
Result
Expected band: 17.9 Kb and 4.9 Kb (EcoRI-HF) and 11.5 Kb, 8.5 Kb and 2.6 Kb (PvuI)
Colony 7 is the only colony which is positiv for all restriction digests.
- => Electroporate plasmid together with DelRest plasmid, as well as the Methylmanolyl-CoA plasmid.
Sequencing of D. acidovorans
PCR Conditions DA.W19.A
For being sure that the mutation at the beginning of DelH is not some mistake in the sequenced genome we amplify this part with one primer binding the delG (FS35) and the screening primer binding at the beginning of DelH (DN07) and let it sequence with GATC.
| Template | 1 µl glycerol stock of D. acidovorans | 1 µl glycerol stock of D. acidovorans |
|---|---|---|
| Expected length [Kb] | 6.5 | 6.5 |
| Named | TD | 2step |
| Primer fw 10 µM | 2 µl FS35 | 2 µl FS35 |
| Primer rev 10 µM | 2 µl DN07 | 2 µl DN07 |
| Phusion Flash (2x) | 10 µl | 10 µl |
| ddH2O | 4 µl | 4 µl |
| DMSO | 1 µl | 1 µl |
| Cycles | Temperature A [°C] | Time [s] | Temperature B[°C] | Time [s] |
|---|---|---|---|---|
| 1 | 98 | 10 | 98 | 10 |
| 12 | 98 | 1 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | - | - | |
| 72 | 130 | 72 | 120 | |
| 18 | 98 | 1 | 98 | 1 |
| 66 | 5 | - | - | |
| 72 | 130 | 72 | 120 | |
| 1 | 72 | 10 min | 72 | 10 min |
| 1 | 12 | inf | 12 | inf |
Result
Expected band: 6.5 Kb
Gel shows expected band.
- => Fragments were cut but not extracted, because side bands were cut too.
- => Run again PCR with 72.3 °C annealing temperature.
PCR Conditions DA.W19.B
| Template | 1 µl glycerol stock of D. acidovorans |
|---|---|
| Expected length [Kb] | 6.5 |
| Named | FS35-DN07 |
| Primer fw 10 µM | 2 µl FS35 |
| Primer rev 10 µM | 2 µl DN07 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 4 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Result
Expected band: 6.5 Kb
Gel shows expected band at ~7 Kb.
- => The fragment was cut and gel purified. Nevertheless the bands were rather weak, so the PCR was repeated.
PCR Conditions DA.W19.C
| Template | 1 µl glycerol stock of D. acidovorans |
|---|---|
| Expected length [Kb] | 6.5 |
| Named | FS35-DN07 |
| Primer fw 10 µM | 2 µl FS35 |
| Primer rev 10 µM | 2 µl DN07 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 4 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Result
Expected band: 6.5 Kb
Gel shows expected fragment.
- => The fragment was cut and gel purified.
- => Send in for sequencing with DN07.
Amplification of 1
09-09 - 15-09-13
Characterization of DelH Plasmid pHM04 30-08 Clones 4, 7, 15
Colony-PCR Conditions CP.W20.A
Because the sequenced DelH-colonies 4, 7, 15 had different kinds of mutations (Deletion of one basepair but also an insertion of a whole sequence part => results are shwon in week 19) we made a new screening PCR of 30 new picked colonies named 31-60 of the plate 2 of the electroporated cells realized in week 19.
There were 30 colonies screened.
| Template | 30x 1 PC of S-A |
| Expected length [bp] | 663 |
| Named | 31-60 |
| Primer fw 10 µM | 30x 2 µl VF2 |
| Primer rev 10 µM | 30x 2 µl DN07 |
| iTaq Polymeras (2x) | 30x 10 µl |
| ddH2O | 30x 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Colonies 33, 39, 40, 46, 51, 58 showed band.
- => Send in for sequencing after isopropanol ethanol purification.
Sequencing
The colonies 33, 39, 40, 46, 51, 58 were sent in MWG for sequencing. There for we prepared 15 µl of the plasmid (midiprep) with a concentration of 50-100 ng/µl and add 2 µl DN07 primer (10µM).
Result
| Colony | Alignment File | Conclusion |
|---|---|---|
| H33 | sequencing insufficient | |
| H39 | sequencing insufficient | |
| H40 | sequencing insufficient | |
| H46 | sequencing insufficient | |
| H51v | sequencing insufficient | |
| H58 | File:Heidelberg Sequencing Result pHM04 DN07 colony H58 DN07.clustal.txt | insertion in the primer region of DelH-backbone (pHM04) |
Colony-PCR Conditions CP.W20.B
New picked olonies 2x 95 well plate
| Template | 95x 1 µl of colony |
| Expected length [bp] | 663 |
| Named | I 1A - I 12H , II 1A - II 12H |
| Primer fw 10 µM | 105x 2 µl VF2 |
| Primer rev 10 µM | 105x 2 µl DN07 |
| iTaq Polymerase (2x) | 105x 10 µl |
| ddH2O | 105x 5 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Colonies collected in table below show a definit result.
- => Add medium and perform a mediprep the next day for send in for sequencing:
| Colony of plate I | Colony of plate II |
|---|---|
| 1F | 2D |
| 1G | 2C |
| 2A | 3G |
| 2H | 4A |
| 3B | 4E |
| 4C | 7C |
| 4E | 7G |
| 4F | 8B |
| 4H | 10E |
| 5E | 10H |
| 5F | 11A |
| 5G | |
| (5H) | |
| 6B | |
| 6C | |
| 6D | |
| 7H | |
| 8B | |
| 8D | |
| 8F | |
| 8G | |
| 9B | |
| 9C | |
| 9F | |
| 10B | |
| 10C | |
| 10G | |
| 11F | |
| 12G |
- => The next step is a test restriction digest with PvuI
Test Restriction Digest
We performed a test digest with PvuI-HF of the screened samples which showed a postive band at 663 bp in the screening PCR. Therefore we prepared a Master Mix with:
| Enzyme | CutSmartBuffer [µl] | ddH2O [µl] | Total amount [µl] |
|---|---|---|---|
| 45x 0.5 µl = 22.5 µl | 45x 2 µl = 90 µl | 45x 16.5 µl = 742.5 µl | 45x 19 µl = 855 µl |
- Incubation time: 1:20 h at 37 °C
Result
Expected bands: 11.5 Kb, 8.5 Kb and 2.6 Kb.
Some colonies showed the expected bands (see table below).
- => These are sent in for sequencing after a miniprep.
The table below shows the result of the restriction digest of different colonies containing the pHM04 plasmid. The restriction digest was positive if the expected bands were pesent.
| Colony of plate I | Show expected bands | Colony of plate II | Show expected bands |
|---|---|---|---|
| 1F | - | 2D | - |
| 1G | - | 2C | - |
| 2A | - | 3G | + |
| 2H | - | 4A | - |
| 3B | - | 4E | + |
| 4C | - | 7C | - |
| 4E | + | 7G | + |
| 4F | - | 8B | + |
| 4H | + | 10E | - |
| 5E | - | 10H | - |
| 5F | - | 11A | - |
| 5G | - | - | - |
| (5H) | - | - | - |
| 6B | + | - | - |
| 6C | + | - | - |
| 6D | + | - | - |
| 7H | + | - | - |
| 8B | - | - | - |
| 8D | - | - | - |
| 8F | - | - | - |
| 8G | + | - | - |
| 9B | - | - | - |
| 9C | - | - | - |
| 9F | - | - | - |
| 10B | + | - | - |
| 10C | - | - | - |
| 10G | + | - | - |
| 11F | + | - | - |
| 12G | - | - | - |
| Colony | Concentration ng/µl |
|---|---|
| I 4E | 1597 |
| I 4H | 495 |
| I 6B | 255 |
| I 6C | 2445 |
| I 6D | 2101 |
| I 7H | 1539 |
| I 8G | 1589 |
| I 10B | 1359 |
| I 10G | 1863 |
| I 11F | 1886 |
| II 3G | 564 |
| II 4E | 2107 |
| II 7G | 1913 |
| II 8B | 955 |
Sequencing
| Colony of plate I | Concentration ng/µl | Amount for sequencing [µl] | ddH2O [µl] | Primer (DN07 [10 µM]) added [µl] |
|---|---|---|---|---|
| I 4E | 1597 | 1 | 14 | 2 |
| I 4H | 495 | 1.5 | 13.5 | 2 |
| I 6B | 255 | 5 | 10 | 2 |
| I 6C | 2445 | 0.5 | 14.5 | 2 |
| I 6D | 2101 | 0.5 | 14.5 | 2 |
| I 7H | 1539 | 1 | 14 | 2 |
| I 8G | 1589 | 1 | 14 | 2 |
Result
| Colony | Sequencing | Notes | Alignment File |
|---|---|---|---|
| I 4E | - | (Deletion of G in coding sequence) | File:Heidelberg Sequencing Result pHM04 DN07 colony I4E.clustal.txt |
| I 4H | - | (Deletion of G in coding sequence) | File:Heidelberg Sequencing Result pHM04 DN07 colony I4H.clustal.txt |
| I 6B | + ? | a deletion in the RBS | File:Heidelberg Sequencing Result pHM04 DN07 colony I6B.clustal.txt File:Heidelberg Sequencing Result pHM04 VF2 colony 6B VF2 VF 2.clustal.txt |
| I 6C | - | (9 deletion) | File:Heidelberg Sequencing Result pHM04 DN07 colony I6C.clustal.txt |
| I 6D | - | Deletion | File:Heidelberg Sequencing Result pHM04 DN07 colony I6D.clustal.txt |
| I 7H | - | Deletion | File:Heidelberg Sequencing Result pHM04 DN07 colony I7H.clustal.txt |
| I 8G | - | Deletion | File:Heidelberg Sequencing Result pHM04 DN07 colony I8G.clustal.txt |
| I 8B | - | Deletion of G in coding sequence | discarded |
| I 10B | - | Deletion of G in coding sequence | discarded |
| I 10G | - | Insertion of G in coding sequence | discarded |
| I 11F | - | Deletion of G in coding sequence | discarded |
| II 3G | - | G Deletion of the ATG (start-codon) and 3 bp later deletion of C | discarded |
| II 4E | - | Deletion of G in coding sequence | discarded |
| II 7G | - | G Deletion of the ATG (start-codon) and 3 bp later deletion of C | discarded |
Because the colony 6B is possibly positive, the next steps are:
- 1. SDS-PAGE
- 2. Sequencing over gibson-assembled parts
- 3. Triple electroporation with the plasmig of Del-rest and MalonylCoA (see lab book Delftibactin)
Amplification of Backbone pSB6A1
PCR Conditions BB.W20.A
| Reagent | BB pSB6A1 | BB pSB6A1 |
|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 |
| Template | 0.5 µl pSB6A1 30-8 | 0.5 µl pSB6A1 30-8 |
| Primer 10 µM fw | 2 µl HM_17 | 2 µl HM_17 |
| Primer 10 µM rev | 2 µl FS_77 | 2 µl FS_77 |
| Phusion Flash Master Mix (2x) | 10 µl | 10 µl |
| DMSO | 1 µl | 1 µl |
| ddH2O | 4.5 µl | 4.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68↓ | 5 | |
| 72 | 3:00 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 3:00 min | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Result
Expected band: 4.4 Kb
Very weak amplification, Primer-Dimers/Oligomers, carry over of Backbone pSB6A1 (including mRFP and therefore creating a double band, since template is 700bp longer than expected PCR product).
- => Repeat PCR with less stringent conditions to increase yield until unexpected bands appear or yield is high enough and reduce amount of template DNA (use 0.5 µL of a 1:10 delution)
- =>Furthermore, elongation time was reduced, as template is only about 4.4 Kb long instead of the putative 7 Kb.
PCR Conditions BB.W20.B
| Reagent | BB pSB6A1 | BB pSB6A1 |
|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 |
| Template | 0.5 µl pSB6A1 (3 ng/µL) | 0.5 µl pSB6A1 (3 ng/µL) |
| Primer 10 µM fw | 2 µl HM_17 | 2 µl HM_17 |
| Primer 10 µM rev | 2 µl FS_77 | 2 µl FS_77 |
| Phusion Flash Master Mix (2x) | 10 µl | 10 µl |
| DMSO | 1 µl | 1 µl |
| ddH2O | 4.5 µl | 4.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66↓ | 5 | |
| 72 | 80 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Result
Expected band: 4.4 Kb

l1:2 log ladder,l2:BB amplified HM17 and HM20 (66°C),l3:BB amplified HM17 and FS77 (66°C),l4:BB amplified HM17 and HM20 (58°C), l5:BB amplified HM17 and FS77 (58°C) - l4-5 show the expected band at 4.4 KB
Gel shows best amplification of fragment using annealing temperature at 58°C.
- => Repetition of PCR at a constant annealing temperature of 58°C.
PCR Conditions BB.W20.C
Performed 6x PCR with FS77 & 6x PCR with HM20, so we have enough yield for the Gibson assembly.
| Reagent | BB pSB6A1 | BB pSB6A1 |
|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 |
| Template | 0.5 µl pSB6A1 (3 ng/µL) | 0.5 µl pSB6A1 (3 ng/µL) |
| Primer 10 µM fw | 2 µl HM_17 | 2 µl HM_17 |
| Primer 10 µM rev | 2 µl FS_77 | 2 µl FS_77 |
| Phusion Flash Master Mix (2x) | 10 µl | 10 µl |
| DMSO | 1 µl | 1 µl |
| ddH2O | 4.5 µl | 4.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Result
Expected band: 4.4 Kb
Gel does not show the expected bands.
- => What happened? Why is it not reproducible? Run a PCR with other conditions.
PCR Conditions BB.W20.D
| Reagent | BB pSB6A1 | BB pSB6A1 |
|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 |
| Template | 1 µl pSB6A1 (3 ng/µL) | 1 µl pSB6A1 (3 ng/µL) |
| Primer 10 µM fw | 2 µl HM_17 | 2 µl HM_17 |
| Primer 10 µM rev | 2 µl FS_77 | 2 µl FS_77 |
| Phusion Flash Master Mix (2x) | 10 µl | 10 µl |
| ddH2O | 5.0 µl | 5.0 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 60↓ | 5 | |
| 72 | 80 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Result
Expected band: 4.4. Kb
Gel shows again no amplification of backbone.
- => Further optimize PCR conditions.
PCR Conditions BB.W20.E
| Reagent | BB pSB6A1 | BB pSB6A1 |
|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 |
| Template | 0.5 µl pSB6A1 (3 ng/µL) | 0.5 µl pSB6A1 (3 ng/µL) |
| Primer 10 µM fw | 1 µl HM_17 | 1 µl HM_17 |
| Primer 10 µM rev | 1 µl FS_77 | 1 µl FS_77 |
| Phusion Flash Master Mix (2x) | 10 µl | 10 µl |
| DMSO | 1 | 1 |
| ddH2O | 6.5 µl | 6.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66↓ | 5 | |
| 72 | 1:20 min | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Result
Expected band: 4.4 Kb
Gel shows amplification using new HM17.
- => Fragments were cut and gel extracted.
PCR Conditions BB.W20.F
Run 6x PCR with HM20 and 6x PCR with FS77 for higher yield
| Reagent | BB pSB6A1 | BB pSB6A1 |
|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 |
| Template | 0.5 µl pSB6A1 (3 ng/µL) | 0.5 µl pSB6A1 (3 ng/µL) |
| Primer 10 µM fw | 1 µl HM_17 | 1 µl HM_17 |
| Primer 10 µM rev | 1 µl FS_77 | 1 µl FS_77 |
| Phusion Master Mix (2x) | 10 µl | 10 µl |
| DMSO | 1 | 1 |
| ddH2O | 6.5 µl | 6.5 µl |
| Cycles | Temperature [°C] | Time |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
The PCR was direcly digested with DpnI afterwards.
Restriction Digest with DpnI
- 10 of the 12 PCRs (5x FS & 5x HM) were cut with DpnI. There we used the exact PCR product as generated earlier.
- Incubated at 37°C ON
- The following table presents the amount for all reactions
| Sample | Buffer | Enzyme | ddH2O |
|---|---|---|---|
| 10x 20 µl PCR | 10x 2.5 µl CutSmart Buffer | 2.5 µl DpnI | 23 µl |
Afterwards, a 0.8% gel was run for 1 h at 100 V and a gel-purification was performed with the Qiagen Gel Extraction Kit
Result
Expected band: 4.4 Kb
Gel shows expected bands.
- => Fragments were gel extracted.
| Sample | DpnI digested | Concentration [ng/µl] |
|---|---|---|
| BB 16 (amplified with FS77) | yes | 6.2 |
| BB 16 (amplified with HM20) | yes | 6.6 |
Generation of DelH Plasmid pHM04 15-09
Gibson Assembly
| Fragment | Concentration [ng/µl] | Amount with BB16 (FS77) [µl] | Amount with BB16 (HM20) [µl] |
|---|---|---|---|
| DN11-FS66 | 172.5 | 0.78 | 0.81 |
| FS67-FS68 | 120.7 | 0.95 | 0.99 |
| FS69-FS70 | 211.3 | 0.94 | 0.98 |
| FS71-HM08 | 146 | 0.55 | 0.57 |
| BB16 amplified with FS77 | 6.2 | 6.78 | - |
| BB16 amplified with HM20 | 6.6 | - | 6.64 |
- Incubate for 1h at 50°C
Electroporation
- Afterwards, 5 µl of each Gibson assembly were taken out and 10 µl ddH2O was added
- With 10 µl of the Gibson assembly, isopropanol purification was performed
- 6 different electroporations were performed (see scheme below)
| Electroporation name | Inserted amount of Gibson Assembly | Plated out on agar plates |
|---|---|---|
| HM20 1 | 1 µl (Gibson + ddH2O) | * 10 µl * rest |
| HM20 14 | 14 µl (Gibson + ddH2O) | * 10 µl * rest |
| HM20 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
| FS77 1 | 1 µl (Gibson + ddH2O) | * 10 µl * rest |
| FS77 14 | 14 µl (Gibson + ddH2O) | * 10 µl * rest |
| FS77 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
Amplification of 1
16-09 - 22-09-13
Generation of DelH Plasmid pHM04 15-09
Colony-PCR Conditions CP.W21.A
| Template | 96x 1 picked colony |
| Expected length [bp] | 663 |
| Named | 1A - 12H |
| Primer fw 10 µM | 100x 2 µl VF2 |
| Primer rev 10 µM | 100x 2 µl DN07 |
| iTaq Polymerase (2x) | 100x 10 µl |
| ddH2O | 96x 6 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
The following colonies seem positive, because they show the expected band at 663 bp.
| Picture | Positive colonies |
|---|---|
| 21.1 | 1B, 1C, 1E, 1F, 1H, 2A, 2B, 2D, 2E, 2F, 3A, 3B, 3C, 3D, 3F, 3G |
| 21.2 | 4A, 4C, 4D, 4G, 5A, 5B, 5C, 5D, 5E, 6A, 6B, 6C, 6E, 6F |
| 22.3 | 7A, 7B, 7C, 7D, 8A, 8B, 8C, 9A, 9E, 9F |
| 22.4 | 11 A, 11E, 11G, 11H, 12D, 12E |
Test Restriction Digest
24 of these positive screened colonies were minipreped and digested with PvuI
| Sample | CutSmartBuffer | PvuI |
|---|---|---|
| 8.5 µl of colonies | 1 µl | 0.5 µl |
Another restriction digest was performed. Colony 7D was digested with EcoRI-HF
| Sample | CutSmartBuffer | EcoRI-HF |
|---|---|---|
| 8.5 µl of colony 7D | 1 µl | 0.5 µl |
Result
Expected bands: 11,621, 8,690 and 2,685 bp (PvuI) and 17,801 and 5,105 bp (EcoRI)
None of the analyzed clones shows correct band pattern.
- => Clones are discarded.
Amplification of Backbone
New Template
Because the backbone amplification yield from pSB6A1 + BBa_J04450 from Spring distribution 2012 (plate 1, well K1) was so low and the screening did not result in positive restriction digests with PvuI or EcoRI, we go a step back and transform E.coli ToP10 with a new pSB6A1 part from the 2013 distribution. Therefore, we transform on the one hand with the part on plate 2 well 2L and on the other hand with the part from the plate 5 well 1K from the Spring 1023 diestricbution.
Aditionally we ran a PCR directly from the two wells with the backbone primers (HM20, HM17 & FS77).
PCR Conditions BB.W21.A
Amplification of pSB6A1 directly from the parts registry
1x PCR with HM_20 and 1x PCR with FS_77
| Reagent | BB pSB6A1 | BB pSB6A1 | BB pSB6A1 | BB pSB6A1 |
|---|---|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 | 4.4 | 4.4 |
| Template | 1 µl pSB6A1 plate 5, well K1 | 1 µl pSB6A plate 2, well L2 | 1 µl pSB6A1 plate 5, well K1 | 1 µl pSB6A plate 2, well L2 |
| Primer 10 µM fw | 1 µl HM_17 | 1 µl HM_17 | 1 µl HM_17 | 1 µl HM_17 |
| Primer 10 µM rev | 1 µl FS_77 | 1 µl FS_77 | 1 µl HM_20 | 1 µl HM_20 |
| Phusion Flash Master Mix (2x) | 10 | 10 | 10 | 10 |
| DMSO | 1 | 1 | 1 | 1 |
| ddH2O | 6 | 6 | 6 | 6 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Result
Expected band: 4. Kb
Amplification worked with the template pSB6A1 from the biobrick distribution 2013 plate 2, well L2.
- => The fragment was gel extracted with QIAquick gel extraction kit, digested with DpnI, and gel extracted again, resulting in final concentrations of 10.7 ng/µl for FS_77 and 4.8 ng/µL for HM_20
- => PCR will be repeated with several samples from the colonies transformed with pSB6A1 from the biobrick distribution 2013 plate 2, well L2
PCR Conditions BB.W21.A
Amplification of backbone fragment pSB6A1 from colonies of DH10ß, transformed with pSB6A1 from the biobrick distribution 2013 plate 2, well L2
4x PCR with HM_20 and 4x PCR with FS_77
| Reagent | BB pSB6A1 | BB pSB6A1 |
|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 |
| Template | colony of DH10B transformed with pSB6A1 plate 5, well K1 | colony of DH10B transformed with pSB6A1 plate 5, well K1 |
| Primer 10 µM fw | 1 µl HM_17 | 1 µl HM_17 |
| Primer 10 µM rev | 1 µl FS_77 | 1 µl FS_77 |
| Phusion Master Mix (2x) | 10 | 10 |
| DMSO | 1 | 1 |
| ddH2O | 6 | 6 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Result
Expected band: 4.4 Kb
Gel shows amplification of fragment.
- => Fragment was cut and gel extracted, then DpnI digested and gel purified again.
Generation of DelH Plasmid pHM04 18-09
Gibson Assembly
| Fragment | Concentration [ng/µl] | Amount with BB16 (FS77) [µl] | Amount with BB16 (HM20) [µl] |
|---|---|---|---|
| DN11-FS66 | 172.5 | 1.77 | 1.84 |
| FS67-FS68 | 120.7 | 2.15 | 2.24 |
| FS69-FS70 | 211.3 | 2.12 | 2.21 |
| FS71-HM08 | 146 | 1.24 | 1.30 |
| BB16 amplified with FS77 | 43.1 | 2.72 | - |
| BB16 amplified with HM20 | 35.8 | - | 2.42 |
- Incubate for 1 h at 50°C
Electroporation
- Afterwards 3 µl of each Gibson assembly were taken and 2 µl ddH2O were added
- With 17 µl of the Gibson assembly, isopropanol purification was performed, DNA was eluted in 20 µl ddH2O
- 6 different electroporations in E.coli BL21 DE3 were performed and grown at RT (see scheme below)
| Electroporation name | Inserted amount of Gibson Assembly | Plated out on agar plates |
|---|---|---|
| HM20 1 | 3 µl (Gibson + ddH2O) | * 10 µl * rest |
| HM20 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
| FS77 1 | 1 µl (Gibson + ddH2O) | * 10 µl * rest |
| FS77 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
Colony-PCR CP.W21.A
| Template | 96x 1 picked colony |
| Expected length [bp] | 663 |
| Named | 1A - 12H DelH I |
| Primer fw 10 µM | 100x 2 µl VF2 |
| Primer rev 10 µM | 100x 2 µl DN07 |
| One-Taq Polymerase (2x) | 100x 10 µl |
| ddH2O | 96x 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
The following colonies seem positive, because they show the expected band at 663 bp
| Picture | Positive colonies |
|---|---|
| 21.12 | - |
| 21.13 | C3, C5, C7, C11, C12, D1, D2, D3, D5, D6, D7, D8, D9, D12 |
| 21.14 | E2, E9, F4, F11 |
| 21.15 | G3, G6, G11, G12, H6, H8, H11 |
Colony-PCR CP.W21.B
| Template | 96x 1 picked colony |
| Expected length [bp] | 663 |
| Named | 1A - 12H, DelH II |
| Primer fw 10 µM | 100x 2 µl VF2 |
| Primer rev 10 µM | 100x 2 µl DN07 |
| One-Taq Polymerase (2x) | 100x 10 µl |
| ddH2O | 96x 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
The following colonies seem positive, because they show the expected band at 663 bp
| Picture | Positive colonies |
|---|---|
| 21.16 | A6, A7, A10, B2, B6 |
| 21.17 | C2, C4, C6, C8, C9, C10, D2, D3, D8, D9 |
| 21.18 | E7, E8, E9, E10, F6 |
| 21.19 | G1, G3, G6, G7, G9, G10, H2, H3, H8, H11 |
Test Restriction Digest
The positive screened colonies were digested with PvuI.
| Template DNA | CutSmart Buffer | PvuI | ddH2O | Total Volume |
|---|---|---|---|---|
| 8.5 µl 1 µl | 0.5 µl | - | 10 µl |
Result
Expected bands: 11,621, 8,628 & 2,685 bp
The following colonies show the correct pattern:
| Colonies | Positive in Digest | Concentration in Midiprep [ng/µl] |
|---|---|---|
| I C5 | + | 454 |
| I C7 | + | 1312 |
| I C12 | + | 5995 |
| I G11 | + | 3887 |
| II A6 | + | 1017 |
| II D2 | + | 926 |
Sequencing
Clones C5, C7, C12, G11, II A6 and II D2 were send for sequencing with VF2.
Result
| Colonies | Alignment File |
|---|---|
| I C5 | File:Heidelberg Sequencing Result pHM04 DN07 colony 05 old.clustal.txt |
| I C7 | sequencing insufficient |
| I C12 | sequencing insufficient |
| I G11 | discarded |
| II A6 | discarded |
| II D2 | discarded |
Amplification of Backbone
PCR Conditions BB.W21.B
Amplification of backbone parts registry well 2L, transformed 19-09 using HPLC purified HM11
| Reagent | HM11 |
|---|---|
| Template | 1 µl of minipreped pSB6A1 (2L) |
| Primer fw 10 µM | 2 µl HM11 |
| Primer rev 10 µM | 2 µl HM17 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0,5°C) | 5 | |
| 72 | 90 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 90 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 4.4 Kb
Gel does not show amplified backbone fragment.
- => Further optimize PCR conditions.
PCR Conditions BB.W21.C
| Reagent | HM11 |
|---|---|
| Template | 1 µl of minipreped pSB6A1 (2L) |
| Primer fw 10 µM | 2 µl HM11 |
| Primer rev 10 µM | 2 µl HM17 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 90 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: ~ 4.4 Kb
Gel shows strong and specific amplification of backbone.
- => Fragments were cut and gel isolated.
Generation of DelH Plasmid pHM04 20-09
Gibson Assembly
| Fragment | Concentration [ng/µl] | Amount with BB16 (FS77) [µl] | Amount with BB16 (HM20) [µl] |
|---|---|---|---|
| DN11-FS66 | 172.5 | 1.77 | 1.84 |
| FS67-FS68 | 120.7 | 2.15 | 2.24 |
| FS69-FS70 | 211.3 | 2.12 | 2.21 |
| FS71-HM08 | 146 | 1.24 | 1.30 |
| BB16 amplified with FS77 | 43.1 | 2.72 | - |
| BB16 amplified with HM20 | 35.8 | - | 2.42 |
- Incubate for 1 h at 50°C
Electroporation
- Afterwards 3 µl of each Gibson assembly were taken and 2 µl ddH2O were added
- With 17 µl of the Gibson assembly, isopropanol purification was performed, DNA was eluted in 20 µl ddH2O
- 6 different electroporations in E.coli DH10ß were performed and grown at RT (see scheme below)
| Electroporation name | Inserted amount of Gibson Assembly | Plated out on agar plates |
|---|---|---|
| HM20 1 | 3 µl (Gibson + ddH2O) | * 10 µl * rest |
| HM20 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
| FS77 1 | 1 µl (Gibson + ddH2O) | * 10 µl * rest |
| FS77 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
New HPLC Purified Primers
As pointed out before, we suspect DelH to be toxic for E. coli and therefore, only colinies containing mutated constructs survive. To avoid this, we ordered new shorter and HPLC purified primers for the assembly of pHM04.

| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| HM_20:BB_HPLC_rev | 11-09-2013 | HPLC version of HM11 Gibson-Primer rev, amplify the Backbone with overlap with the RBS and the lacI-promotor and it creates and overlap to the start of DelH | GATTTGGCGCAGGCGGCCACGGTCCATctagtatttctcctctttc |
| FS_77:BB_HPLC_rev | 11-09-2013 | Gibson-Primer rev, amplify the Backbone with overlap with the RBS and the lacI-promotor and it creates and overlap to the start of DelH | GCGATTTGGCGCAGGCGGCCACGGTCCATCTAGTATTTCTCCTCTTTC |
Mutagenesis of DelH clones I 6B and 15
Strategy
As our clones which have the DelH construct keep on having mutations and deletions we decided to fix this problem by carrying out site-directed mutagenesis. The targeted mutations are the deletion of parts of the ribsome binding site in clone I6B and the deletion at the beginning of DelH in clone 15. For this we make use of two restriction enzymes (MfeI & XbaI) which both only cut in the construct once. By this we excise the fragment with the mutation. The larger fragment (17.3 kbp) without mutation is kept.
Furthermore we ordered four new primers. The smaller fragment is then PCR amplified with those primers in two fragments. Primer HM22 and HM23 cure the mutations. With those primers also recognitions sites for the restriction enzyme BbsI are introduced, which is needed for religation of the two fragments. BbsI cuts two basepair after its recognition site.
HM21 and HM24 are the corresponding primer. They introduce XbaI and MfeI sites as well as in each on BbsI recognition site. Those are needed for religation.
In the end the two fragments are both digested with BbsI. In the adjacent ligation the two fragments can ligate together due to the BbsI recognition sites. Furthermore BbsI opens the introduced MfeI and XbaI sites through which the two fragments can ligate with the digested 17.3 kb fragment.
However the XbaI site is not introduced correctly again. Therefore we can test if the mutagenesis had been successful by digesting with XbaI. If the site is not present anymore we successfully got rid of the mutations.
New Primers
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| HM21_fw_lacI_BbsI_Xba | 2013-09-15 | Forward primer for cutting out mutated fragment for mutagenesis | TTTTGAAGACAA CTAGGCAATACGCAA |
| HM22_rev_RBS | 2013-09-15 | Reverse Primer in RBS for mutagenesis | TTTTGAAGACAA CTCTTTCTCTAGTATGTGTGAAATTG |
| HM23_fw_RBS | 2013-09-15 | Forward Primer in RBS for mutagenesis | TTTTGAAGACAA AGAGGAGAAATACTAGATGGACCGTGGC |
| HM24_rev_BbsI_MfeI | 2013-09-15 | Reverse primer for cutting out mutated fragment for mutagenesis | TTTTGAAGACAA AATTGGACAGCGCGGCATGCCGGTTG |
Purification of Midiprep of Clone I6B
As can be seen in figure 21.23, the midi-prep of colony I 6B is contaminated. Thus, we tried to purify only the largest band by gel extraction with the QIAEX II Gel Extraction Kit (Qiagen). The procedure was as following:
- Application of 2 µl of midiprep on gel (4596.7 ng/µl --> ~10 µg)
- Excision of band at about 17.4 kb
- Gel extraction of DNA with QIAEX II Gel Extraction Kit (Qiagen)
Result
- The concentration of the purified fragment was only 4.7 ng/µl after extraction.
- => We are trying to elute again.
- The second elution did not work either
Amplification of HM23 to HM24
1x 20 µl for each colony
| Template | 0.2 µl of either midiprep colony I6B or 15 |
|---|---|
| Expected length [kbp] | 5.3 |
| Named | I6B or 15 HM23-HM24 |
| Primer fw 10µM | 2 µl HM23 |
| Primer rev 10 µM | 2 µl HM24 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 s |
| 12 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | |
| 72 | 1:50 min | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 1:50 min | |
| 1 | 72 | 7 min |
| 1 | 12 | inf |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 69 | 5 | |
| 72 | 1:50 min | |
| 1 | 72 | 7 min |
| 1 | 12 | inf |
| Template | 0.2 µl of either midiprep colony I6B or 15 |
|---|---|
| Expected length [kbp] | 5.3 |
| Named | I6B or 15 HM23-HM24 |
| Primer fw 10µM | 2 µl HM23 |
| Primer rev 10 µM | 2 µl HM24 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 4 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 s |
| 30 | 98 | 1 s |
| 72 | 1:40 min | |
| 1 | 72 | 7 min |
| 1 | 12 | inf |
Result
Gels for amplification of HM23 to HM24
Amplification of HM21 to HM22
| Template | 0.2 µl of either midiprep colony I6B or 15 |
|---|---|
| Expected length [kbp] | 200 bp |
| Named | I6B or 15 HM21-HM22 |
| Primer fw 10µM | 2 µl HM21 |
| Primer rev 10µM | 2 µl HM22 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 63 | 5 | |
| 72 | 10 | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
Result
Gels for amplification of HM21 to HM22
Restriction Digest of pHM04 (I6B) with XbaI and MfeI
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B or 15 | 0.5 |
| Cutsmart buffer | 2 |
| XbaI | 1.5 |
| MfeI-HF | 1.5 |
| ddH2O | 14.5 |
- Incubation for 2 h at 37°C
Result
- Expected band pattern: 17.3 kbp, 5.3 kbp
Gels for restriction Digest of pHM04 (I6B) with XbaI and MfeI
- The MfeI already expired in 2011
- => We will test digest the construct again with a new enzyme
Restriction Digest of pHM04 (I6B) with XbaI and MfeI
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B or 15 | 0.5 |
| Cutsmart buffer | 5 |
| XbaI | 1.5 |
| MfeI-HF | 1.5 |
| ddH2O | 14.5 |
| Incubation for 2 h at 37°C | Expected band pattern: 17.3 kbp, 5.3 kbp |
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B purified 1:10 | 1 |
| Cutsmart buffer | 2 |
| XbaI | 1 |
| MfeI-HF | 1 |
| ddH2O | 15 |
| Incubation at room temperature, over night | Expected band pattern: 17.3 kbp, 5.3 kbp |
Result
Due to the contamination in DelH it was not possible to decide whether the fragment really had been digested. Thus we have to test whether MfeI Cutting site is really present.
Test for MfeI Cutting Site
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B | 0.5 |
| Cutsmart buffer | 2 |
| NotI-HF | 1 |
| ddH2O | 16.5 |
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B | 0.5 |
| Cutsmart buffer | 2 |
| NotI-HF | 1 |
| MfeI-HF | 1 |
| ddH2O | 15.5 |
Incubation at room temperature over night.
--> As the upper band is lower for the digest with both enzymes the MfeI restriction site is apparently present. Therefore the mutagenesis is tried.
Restriction Digest of Fragment HM21-HM22 (I6B) and HM23-HM24 (I6B) with BbsI
PCR fragments: HM21-HM22 (36.6 ng/µl) and HM23-HM24 (32.4 ng/µl)
| Reagent | Volume [µl] |
|---|---|
| PCR fragment | 20 |
| 2.1 buffer | 5 |
| BbsI | 1 |
| ddH2O | 24 |
Ligation of Digested pHM04 (I6B), HM21-HM22 and HM23-HM24
| Reagent | Volume [µl] |
|---|---|
| digested and purified pHM04 (I6B) | 3 |
| digested HM21-HM22 | 0.5 |
| digested HM23-HM24 | 0.5 |
| T4 ligase buffer | 2 |
| T4 ligase | 1 |
| ddH2O | 13 |
Transformation of Ligation
1 µl and 5 µl were transformed in 50 µl electrocompetent DH10β each by electroporation. The cells were recovered in 400 µl SOC media and plated on LB Amp.
Amplification of 1
23-09 - 29-09-13
Mutagenesis of DelH Clone I 6B
Screening-PCR Conditions CP.W22.A
Screening of the 11 colonies, which grew on the agar plate.
| Template | 11x 1 picked colony + positve control (Midiprep I6B) |
| Expected length [bp] | 319 (VFII+HM22); 395 (FS66+SR03) |
| Named | Mut1-11 (VFII+HM22), Mut1-11 (FS66+SR03) |
| Primer fw 10 µM | 2 µl VF2 / FS66 |
| Primer rev 10 µM | 2 µl HM22 / SR03 |
| One-Taq (2x) | 10 µl |
| ddH2O | 6 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 900 |
| 35 | 95 | 30 |
| 55 | 30 | |
| 72 | 25 | |
| 1 | 12 | inf |
Result
Expected bands: 319 bp (HM22-VFII) and 395 bp (FS_66-SR_03)
10 ml of each colony were cultured.
- The screening with primer FS_66 to SR_03 did not work. The positive control was negative.
- The screening with primer VFII to HM22 was positive for 4 colonies (2,3,4,7,8,10)
- These colonies have to be midi-preped and digested to test whether it is the original DelH or the mutated one
Midiprep of Clones 2,3,4,7,8 and 10
- Centrifugation of 10 ml culures at 3750 rpm for 15 min
- Kept pellet an resuspended in 400 µl P1 buffer
- Added 800 µl P2 buffer and incubated for 180
- Neutralized with S3 buffer
- Centrifugated for 15 min at 13 000 rpm
- Kept supernatant and added 50:50 Isopropanol
Test restriction digest of Midipreps with MfeI and XbaI
Expected band: 22.9 kb
| Reagent | Volume [µl] |
|---|---|
| Midiprep colonies | 0.2 |
| Cutsmart buffer | 2 |
| XbaI | 1.5 |
| MfeI-HF | 1.5 |
| ddH2O | 14.75 |
- Incubation for 2 h at 37°C
Apparently the mutagenesis did not work. The band was too low. Maybe recombination occured.
Sequencing
The Midiprep of clone 2 was send to sequencing to see what had happened during the mutagenesis.
Result
The mutagenesis did not work. Apparently recombination occured. As the result was the same for all the colonies we suspect that repeating the mutagenesis would lead to the same result. Therefore the mutagenesis part is stopped.
SDS-PAGE of Clone C5
Lysis of Cells
- 1 ml over night cultures in LB of electrocompetent BL21-pLys and BL21-pLys with pHM04 (C5)
- Meausured ODs (BL21-pLys = 1.33; BL21-pLys + pHM04= 1.03 )
- Centrifugation at 13000 rpm for 10 min
- Discarded supernatant
- Resuspended cells in 133 µl (BL21-pLys) and 103 µl (BL21-pLys + pHM04) 1x loading buffer (40% ddH2O, 12.5% TrisHCl pH=7.5, 20% Glycerol (50%), 20% SDS (10%), 5% β-Mercaptoethanol, 2.5% Bromophenol blue)
- Boiled samples for 10 min at 98°C
Electrophoresis
- Applied 25 µl of each sample on SDS-polyacrylamide gel
- Run at 100 V for 5 min and afterwards at 140 V for another 50 min
Staining and Destaining Procedure
- Gel was stained in Coomassie Brilliant Blue for 1 h on shaker
- Destaining with destaining buffer (50% ddH2O, 40% Methanol, 10% Acetic acid) until band were visible
Result
As shown in figure 22.3, there is a faint very high band, which is DelH (~600 kDa). Clone C5 expresses DelH despite of the amino acid substitution. Next, we will electroporate DelH C5 with DelRest and pIK8.6 in E. coli BL21 DE3 and analyze Delftibactin expression by Micro-TOF.
Generation of DelH Plasmid pHM04 18-09
Colony-PCR CP.W22.B
| Template | 48x 1 picked colony |
| Expected length [bp] | 663 |
| Named | 1A - 12E, DelH III |
| Primer fw 10 µM | 50x 2 µl VF2 |
| Primer rev 10 µM | 50x 2 µl DN07 |
| One-Taq Polymerase (2x) | 50x 10 µl |
| ddH2O | 48x 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 s | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
- The following colonies seem positive, because they show the expected band at 663 bp
| Picture | Positive colonies |
|---|---|
| 22.1 | A9 |
| 22.2 | D7, D9, D11 |
| 22.3 | E5, E6, E7, F7 |
Test Restriction Digest
The positive screened colonies were digested with PvuI.
| Template DNA | CutSmart Buffer | PvuI | ddH2O | Total Volume |
|---|---|---|---|---|
| 8.5 µl 1 µl | 0.5 µl | - | 10 µl |
Result
Expected bands: 11,621, 8628 & 2,685 bp
The following colonies show the correct pattern:
| Colonies | Positive in Digest | Concentration in Midiprep [ng/µl] |
|---|---|---|
| II A9 | yes | 2,153 |
| III D7 | yes | 3,371 |
| III D9 | yes | 697 |
| III D11 | yes | 3,523 |
| III E6 | yes | 2,745 |
| III E7 | yes | 3,649 |
They were not sequenced due to exclusively found mutations in BL21 DE3 (pLys).
Generation of DelH Plasmid pHM04 20-09
Colony-PCR CP.W22.C
| Template | 48x 1 picked colony |
| Expected length [bp] | 663 |
| Named | 1F - 12H, DelH III |
| Primer fw 10 µM | 50x 2 µl VF2 |
| Primer rev 10 µM | 50x 2 µl DN07 |
| One-Taq Polymerase (2x) | 50x 10 µl |
| ddH2O | 48x 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 s | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
- The following colonies seem positive, because they show the expected band at 663 bp
| Picture | Positive colonies |
|---|---|
| 22.3 | F7, F9, F11 |
| 22.4 | G3, G4, G12, H6, H11 |
Test Restriction Digest
The positive screened colonies were digested with PvuI.
| Template DNA | CutSmart Buffer | PvuI | ddH2O | Total Volume |
|---|---|---|---|---|
| 8.5 µl 1 µl | 0.5 µl | - | 10 µl |
Result
Expected bands: 11,621, 8628 & 2,685 bp
The following colonies show the correct pattern:
| Colonies | Positive in Digest | Concentration in Midiprep [ng/µl] |
|---|---|---|
| III F7 | yes | 3,856 |
| III F9 | yes | 1,283 |
| III F11 | no | 4,507 |
| III G3 | yes | 5,823 |
| III G4 | yes | 3,622 |
| III G12 | no | 3,615 |
| III H6 | no | 3,985 |
| III H11 | no | 3,605 |
Sequencing
Colonies F7, F9, G3 and G4 were send for sequencing with VF2.
Result
Contamination of Cells with Chloramphenicol Resistance
Preparation of Electrocompetent BL21 DE3
Electrocompetent BL21 DE3 were prepared according to the protocol. The following day, the LB Chlor plate was covered with colonies. Since BL21 are in contrast to BL21 pLys not ntaurally resistant to Chloramphenicol, they are obviously contaminated. In order to check whether this is also caused by an Indigoidine plasmid, extensive evaluation of stocks and transformed cells will be undertaken.
Test for Chloramphenicol Resistance
All stocks of competent cells were streaked on LB Chlor plates and incubated at 37°C ON.
Result
| Cells | Prepared | Growth on Plate |
|---|---|---|
| E.coli DH10ß elec | older | no |
| E.coli DH10ß elec | 21-09 | no |
| E.coli BL21 DE3 chem | original stock | yes |
| E.coli BL21 DE3 elec | older | yes |
| E.coli BL21 DE3 elec | 22-09 | yes |
| E.coli BL21 pLys elec | older | yes |
Thus, the BL21 DE3 we obtained most probably are BL21 DE3 pLys, too.
New BL21 DE3
We obtained new BL21 DE3 from the Russel Lab, which were obtained from NEB directly. Cells were inocculated in 10 ml LB and streaked on LB Chlor plates. The vial was frozen again at -80°C.
Test for Indigoidine Contamination A
All competent cells as well as important transformed cells and recent mini preps of possible DelH plasmids were tested for contamination with an Indigoidine plasmid (which carries chloramphenicol resistance).
| Reagent | Amount [µl] |
|---|---|
| Template | 0.5 or colony |
| RB69 10 µM | 1 |
| VF2 10 µM | 1 |
| OneTaq Master Mix | 5 |
| ddH2O | 2.5 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 53 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
Result
Expected band: 540 bp
The following samples show Indigoidine band:
| Name | Cells | Plasmids | Indigoidine band |
|---|---|---|---|
| 1 | E.coli BL21 DE3 original | - | no |
| 2 | E.coli BL21 DE3 older | - | no |
| 3 | E.coli BL21 DE3 22-09 | - | no |
| 4 | E.coli DH10ß 21-09 LB | - | no |
| 5 | miniprep | DelH C5 | yes |
| 6 | miniprep | DelH C7 | yes |
| 7 | miniprep | DelH C12 | yes |
| 8 | miniprep | DelH G11 | yes |
| 9 | miniprep | DelH IIA6 | yes |
| 10 | miniprep | DelH IID2 | yes |
| 11 | E.coli BL21 DE3 older | DelCluster 25-1B + pIK8.6 | no |
| 12 | E.coli BL21 DE3 older | DelCluster 25-1C + pIK8.6 | no |
| 13 | E.coli BL21 DE3 older | DelCluster 25-2A + pIK8.6 | no |
| 14 | E.coli BL21 DE3 older | DelCluster 25-3A + pIK8.6 | no |
| 15 | E.coli BL21 DE3 older | DelCluster 25-3B + pIK8.6 | no |
| 16 | E.coli BL21 DE3 older | DelCluster 25-4 + pIK8.6 | no |
| 17 | E.coli BL21 DE3 older | DelCluster 25-5 + pIK8.6 | no |
| 18 | E.coli BL21 pLys | - | no |
| 19 | E.coli BL21 pLys | DelH C5 + pIK8.6 | no |
| 20 | E.coli BL21 pLys | DelH C7 + pIK8.6 | no |
| 21 | E.coli BL21 pLys | DelH C12 + pIK8.6 | no |
| 22 | E.coli BL21 pLys | DelH G11 + pIK8.6 | no |
| 23 | E.coli BL21 pLys | DelH IIA6 + pIK8.6 | no |
| 24 | E.coli BL21 pLys | DelH IID2 + pIK8.6 | no |
| 25 | miniprep | pIK2.6 | yes |
| 26 | E.coli DH10ß white colony | DelRest + DelH I6B + pIK2.6 | no |
| 27 | E.coli DH10ß 21-09 ACM | - | no |
| + | miniprep | pRB22.4 | yes |
All stocks are clean, only the DelH minipreps as well as the one from pIK2.6 are positive for Indigoidine. Therefore, we identified the source of contamination for the triple clone, which turned blue. We however cannot explain the positive result for the DelH minipreps, which obviously harbour a second plasmid.
Test for Indigoidine Contamination B
Again, all minipreps which were send for sequencing as well as a positive control a negative control and two recently prepared minipreps from other teams were tested for an Indigoidine contamination
| Reagent | Amount [µl] |
|---|---|
| Template | 0.5 or colony |
| RB69 10 µM | 1 |
| VF2 10 µM | 1 |
| OneTaq Master Mix | 5 |
| ddH2O | 2.5 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 53 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
Result
Expected band: 540 bp
| Lane | Cells originating from | Miniprep | Indigoidine band |
|---|---|---|---|
| upper row | |||
| 2 | E.coli BL21 DE3 22-09 | DelH Plasmid I C5 23-09 | yes |
| 3 | E.coli BL21 DE3 22-09 | DelH Plasmid I C7 23-09 | yes |
| 4 | E.coli BL21 DE3 22-09 | DelH Plasmid I C12 23-09 | yes |
| 5 | E.coli BL21 DE3 22-09 | DelH Plasmid I G11 23-09 | yes |
| 6 | E.coli BL21 DE3 22-09 | DelH Plasmid II A6 23-09 | yes |
| 7 | E.coli BL21 DE3 22-09 | DelH Plasmid II D2 23-09 | yes |
| 8 | E.coli BL21 DE3 22-09 | DelH Plasmid III A9 23-09 | yes |
| 9 | E.coli BL21 DE3 22-09 | DelH Plasmid III D7 23-09 | yes |
| 10 | E.coli BL21 DE3 22-09 | DelH Plasmid III D9 23-09 | yes |
| 11 | E.coli BL21 DE3 22-09 | DelH Plasmid III D11 23-09 | yes |
| 12 | E.coli BL21 DE3 22-09 | DelH Plasmid III E6 23-09 | yes |
| 13 | E.coli BL21 DE3 22-09 | DelH Plasmid III E7 23-09 | yes |
| 14 | E.coli DH10ß older | DelH Plasmid III F6 23-09 | yes |
| 15 | E.coli DH10ß older | DelH Plasmid III F7 23-09 | yes |
| 16 | E.coli DH10ß older | DelH Plasmid III G3 23-09 | yes |
| 17 | E.coli DH10ß older | DelH Plasmid III G4 23-09 | yes |
| 18 | E.coli DH10ß chem | Indigoinide Plasmid pRB22 (positive) | yes |
| lower row | |||
| 2 | E.coli DH10ß chem | Indigoinide Plasmid pRB17 (negative) | yes |
| 3 | E.coli DH10ß chem | Indigoinide Plasmid recent miniprep K1 (negative) | yes |
| 4 | E.coli DH10ß chem | Indigoinide Plasmid recent miniprep K1 (negative) | yes |
| 5 | E.coli DH10ß chem | Indigoinide Plasmid pRB22 (positive) | yes |
Surprisingly, all samples show Indigoidine band at ~540 bp. There is a contamination and we have to identify the source!
Test for Indigoidine Contamination C
Again, all minipreps which were send for sequencing as well as positive and negative controls and recently prepared minipreps from other teams were tested for an Indigoidine contamination. using two different primer pairs. Additionally, fake minipreps were performed to identify DNA contamination of buffers.
| Reagent | Amount [µl] |
|---|---|
| Template | 0.5 or colony |
| RB69 10 µM | 1 |
| VF2 10 µM | 1 |
| OneTaq Master Mix | 5 |
| ddH2O | 2.5 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 53 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
| Reagent | Amount [µl] |
|---|---|
| Template | 0.5 or colony |
| KH05 10 µM | 1 |
| VR 10 µM | 1 |
| OneTaq Master Mix | 5 |
| ddH2O | 2.5 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 53 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
Result
Expected band: ~1 Kb for KH05 and 540 bp for RB69
| Lane | Date | Template | Indigoidine band |
|---|---|---|---|
| KH05 and VR | |||
| 2 | 24-09 (negative) | Fake miniprep: all old | no |
| 3 | 24-09 (negative) | Fake miniprep: P1 aliquot 20-09 | no |
| 4 | 24-09 (negative) | Fake miniprep: fresh P1 | no |
| 5 | 24-09 (negative) | Fake miniprep: fresh P2 | no |
| 6 | 24-09 (negative) | Fake miniprep: P2 aliquot | no |
| 7 | 24-09 (negative) | Fake miniprep: fresh S3 | no |
| 8 | 24-09 (negative) | Fake miniprep: fresh isoprop | no |
| 9 | 21-07 (negative) | pKH1 | no |
| 10 | 21-07 (negative) | pKH1 + new polymerase | yes |
| 11 | 21-07 (negative) | pSB3C5 + new polymerase | yes |
| 12 | - | pRB22 (positive) | yes |
| 13 | 23-09 (negative) | DelH Plasmid I C7 23-09 | yes |
| 14 | 23-09 (negative) | DelH Plasmid I C12 23-09 | yes |
| 15 | 23-09 (negative) | DelH Plasmid I G11 23-09 | yes |
| 16 | 23-09 (negative) | DelH Plasmid II A6 23-09 | yes |
| 17 | 23-09 (negative) | DelH Plasmid II D2 23-09 | yes |
| 18 | 23-09 (negative) | DelH Plasmid III F7 23-09 | yes |
| 19 | 23-09 (negative) | DelH Plasmid III F9 23-09 | yes |
| 20 | 23-09 (negative) | DelH Plasmid III G3 23-09 | yes |
| 21 | 23-09 (negative) | DelH Plasmid III G4 23-09 | yes |
| 22 | 23-09 (negative) | DelH Plasmid I C5 23-09 | yes |
| 23 | E.coli DH10ß culture (negative) | DelH Plasmid II A6 | no |
| 24 | - | - | no |
| Lane | Date | Template | Indigoidine band |
|---|---|---|---|
| RB69 and VF2 | |||
| 2 | 24-09 (negative) | Fake miniprep: all old | yes |
| 3 | 24-09 (negative) | Fake miniprep: P1 aliquot 20-09 | yes |
| 4 | 24-09 (negative) | Fake miniprep: fresh P1 | yes |
| 5 | 24-09 (negative) | Fake miniprep: fresh P2 | yes |
| 6 | 24-09 (negative) | Fake miniprep: P2 aliquot | yes |
| 7 | 24-09 (negative) | Fake miniprep: fresh S3 | yes |
| 8 | 24-09 (negative) | Fake miniprep: fresh isoprop | yes |
| 9 | 04-08 (negative) | 6 maxi HM | no |
| 10 | 15-09 (negative) | pPW06 BCI | no |
| 11 | 19-09 (negative) | ccdB 7 | no |
| 12 | 19-09 (negative) | c.3 7 | no |
| 13 | 19-09 (negative) | c.3 14.B | no |
| 14 | 21-07 (negative) | pKH1 | no |
| 15 | - (positive) | pRB22 | yes |
| 16 | - | - | no |
Amplification with KH05 shows contamination of new polymerase vial (shorter band) as well of DelH minipreps. Interestingly, the tested culture is NOT contaminated.
Amplification of 1
== 30-09 - 06-10-13 ==
Generation of DelH Plasmid pHM04 15-09
Colony-PCR Conditions CP.W23.A
| Template | 23x 1 picked colony, positive control |
| Expected length [bp] | 663 |
| Named | 1A - 12H |
| Primer fw 10 µM | 100x 2 µl VF2 |
| Primer rev 10 µM | 100x 2 µl DN07 |
| iTaq Polymerase (2x) | 100x 10 µl |
| ddH2O | 96x 6 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 ↓ 0.5 | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
[[|200px|thumb|right|Fig.23.1 Colony PCR of Gibson assembled DelH-BB without mRFP 23 colonies]]
Amplification of backbone pSB6A1 for strategy pFS02
| Reagent | Amount |
|---|---|
| Template | 1 µl of minipreped pSB6A1 (2L) |
| Primer fw 10 µM | 2 µl FS78 |
| Primer rev 10 µM | 2 µl HM17 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 1:25 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 1:25 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 58 | 5 | |
| 72 | 1:25 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Results
Amplification of backbone pSB6A1 for strategy pFS02
| Reagent | Amount |
|---|---|
| Template | 1 µl of minipreped pSB6A1 (2L) |
| Primer fw 10 µM | 2 µl FS85 |
| Primer rev 10 µM | 2 µl HM17 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 1:25 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 1:25 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 58 | 5 | |
| 72 | 1:25 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
====Result====
 "
"