Team:Heidelberg/HumanPractice/iGEM42
From 2013.igem.org
| Line 21: | Line 21: | ||
</div> | </div> | ||
<div> | <div> | ||
| - | + | <p style="text-align:justify"><a class="btn btn-lg btn-default" href=http://igem2013.bioquant.uni-heidelberg.de/shiny/iGEMPulse/homepage/index_iGEM42.htmlstyle="position:absolute; vertical-align: middle;">Download Safety Form</a></p> | |
| + | |||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
Revision as of 01:23, 5 October 2013


iGEM 42. The answer to 'almost' everything.
Have you ever easily found the piece of information you wanted from past iGEM competitions? Right! That turned out to be very hard.
Maybe you should try this tool which was developed to do exactly this: Lead you directly to the information you want.
Our motivation
A situation all new iGEM teams have to face in the beginning of their project is the intensive and sometimes frustrating phase of collecting novel ideas that potentially lead to the one and only BioBrick. The new concept should be at best totally independent from earlier iGEM projects yet exciting enough to reach the full attention of the iGEM community. Because we found it very tedious to search manually through every single wiki we thought of other, automated possibilities to make this process more bearable. Intriguingly, latest web-based software tools offer the possibility to strategically search through other web pages and directly ‘scrape out’ the wanted information.
Having this tool at hand we thought of building a web application that systematically integrates and refines information from all existing iGEM wikis. As a reference to the famous book "The Hitchhiker's Guide to the Galaxy" we named this tool iGEM42 as the potential answer to every question a future iGEM veteran could think of. Maybe it is impossible to answer the question in one sentence, what it takes to win the iGEM competition. The contest is integrating so many different aspects of natural sciences and beyond that even experienced iGEM members tend to overlook the little details that could make a difference in the end. Nevertheless we wanted to give it a try and you can now get to know our first implementation of iGEM42 that focuses on a statistical analysis of the characteristics of previous iGEM teams. You can go deeper into the matter and search for the decisive aspects that let a team win or lose. Read on and find out about the technical details of the implementation or directly move to the GUI and get familiar with iGEM42.
Implementing a web scraping algorithm
Since the data available on all the iGEM wikis out there is really massive, a realtime analysis would be very time consuming. So we had to collect the data using a python-based scraping tool crawling though all the team and results overview pages of the last years iGEM teams. This data was then analysed by text mining algorithms like together with the statistical framework R. The analyses includes topic extraction based on key terms in synthetic biology as well as a scoring system for the different medals and awards throughout the history of iGEM.
[1]
For this scoring function, we tried to define significant parameters that should sufficiently characterize a particular iGEM team. Besides the overall number of awards, the number of submitted BioBricks and the number of regional awards we defined our own scoring function returning a single success value for each team. To weight a particular award against another is a delicate issue and highly subjective. However, it is necessary if you, for example, consider the different value of the grand prize compared to a regional special prize. Our solution was an internal poll where the team members were asked to rank every award according to their own opinion. In the future this scoring function could be further generalized by organizing a questionnaire among all iGEM teams participating.
To display the results in an intuitive and appealing way we designed a set of matching shiny applications serving different needs, that can be easily integrated in any webpage. One allows you to investigate the correlation between selected parameters or observe the development of a certain parameter over time. Two other ones can let you know, if your method of choice was already used by another team or if someone worked on your topic before, by simply matching a keyword you typed into the input form.
What is there to be learned?
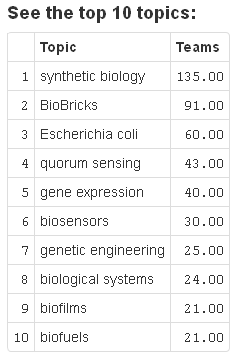
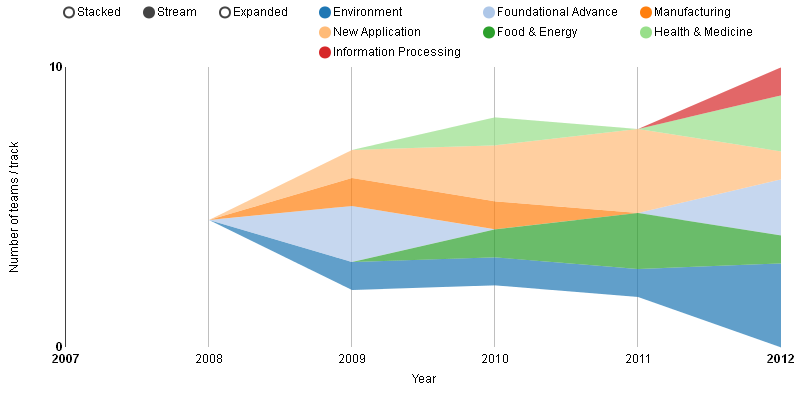
The most interesting findings one can have using iGEM 42 are probably those hidden in the wikis. They can be best accessed via the topics app, because the text mining has a very broad basis of key terms in synthetic biology. The list of the top 10 topics in iGEM competitions is displayed in figure 1. Since biofuels made it on this list one can conclude, that environmental issues are a hot topic in synthetic biology. Another argument for this is, that the environment track is one of those which always contained teams scoring high in the competition. This can be seen in figure 2, which displays a timeline of the teams having a score of 20% or higher in our scoring system, where the color of the area represents the corresponding track. The only track scoring even higher than environment is new application. In contrast to what one might expect foundational advance teams do not tend to score very high. The one track, that never had a high scoring team at all is the software track, which is also due to the separate software division, existing since 2011.
iGEM42 2.0
Until now the result is scraped only out of the wiki abstracts of previous iGEM teams. Of course this approach is very limited however this is planned to be extended by scraping the whole team wikis and running optimised text-mining scripts on the plain text contents. Other features one could include are a histogram-like display of the numerical parameters and displaying more information, as for example the team abstracts, directly within the tool.
1. Oldham, P., Hall, S., & Burton, G. (2012). Synthetic Biology: Mapping the Scientific Landscape. PLoS ONE, 7(4), e34368. doi:10.1371/journal.pone.0034368
 "
"