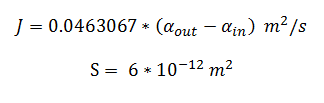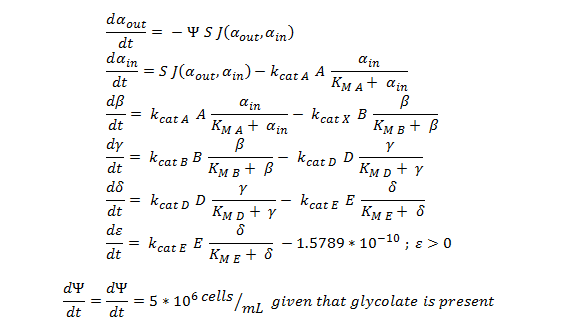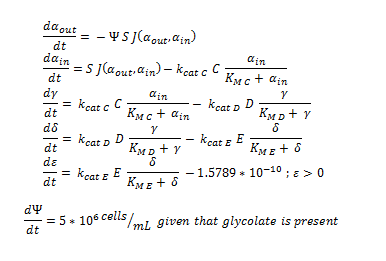Team:SydneyUni Australia/Modelling Output
From 2013.igem.org


Contents |
Running the Model
The model was run using MATLAB through the ODE solver ODE45.
initial conditions':
| Constant | Value | Comment |
|---|---|---|
| KM A | 0.530 mM | From literature [1] |
| kcat A | 3.3 s-1 | From literature [1] |
| KM B | 0.940 mM | From literature [2] |
| kcat B | 0.0871 s-1 | From literature [2] |
| KM C | 7.2 mM | From literature [3] |
| kcat c | 89.8 s-1 | From literature [3] |
| KM D | 0.160 mM | From literature [4] |
| kcat D | 0.600 s-1 | From literature [4] |
| KM E | 20 mM | From literature [5] |
| kcat E | 25.4 s-1 | From literature [5] |
| β, γ, δ & ε | 0 mM | Not naturally present in cells. |
| αout | 1 mM | Initial concentration of DCA in solution (arbitrary) |
| αin | 0.001 mM | Initial concentration of DCA in cell |
| A, B, C, D, E | 25.55 mM | Estimation from literature [11], as described in Validation. |
| 2 x 108 cells / mL | Initial cell concentration which allows appropriate growth |
Physical conditions:
The model assumes that the cells are present in minimal media prior to DCA exposure and that the DCA is instantaneously and evenly present at time = 0 min in a solution of homogeneously mixed cells Many other assumptions have been made in the construction of the model and are outlined in the mathematical model section.
Flux rate:
By using the constants summarised in the previous section the flux, J, took the value (alongside the bacterial surface area, S):
The Non-monooxygenase Pathway
Summary of the ODE:
This system of ODE has been explained and justified in the previous section. It has been converted into MATLAB code in order to run the model, allowing a computer to generate the graphs below.
Raw MATLAB code:
function dy = nop450(t,y)
dy=zeros(7,1);
dy(1)= -y(7)*(6*10^(-12))*0.0463067*(y(1)-y(2));
dy(2)= ((6*10^(-12))*0.0463067*(y(1)-y(2)))-3.3*25.55*(y(2)/(0.53+y(2)));
dy(3)= 3.3*25.55*(y(2)/(0.53+y(2)))-0.0871*25.55*(y(3)/(0.94+y(3)));
dy(4)= .0871*25.55*(y(3)/(0.94+y(3)))- 0.6*25.55*(y(4)/(0.16+y(4)));
dy(5)= 0.6*25.55*(y(4)/(0.16+y(4))) - 25.4*25.55*(y(5)/(20+y(5)));
if y(6) > 2*10^(-10)
dy(6)= 25.4*25.55*(y(5)/(20+y(5))) -1.5789*10^(-10)
else
dy(6) = 25.4*25.55*(y(5)/(20+y(5)))
end
if y(6) > 0.0005
if y(7) > 1.6*10^11
dy(7)=0
else
dy(7) = 5*10^6
end
else
dy(7) = 0
end
end
MATLAB output:
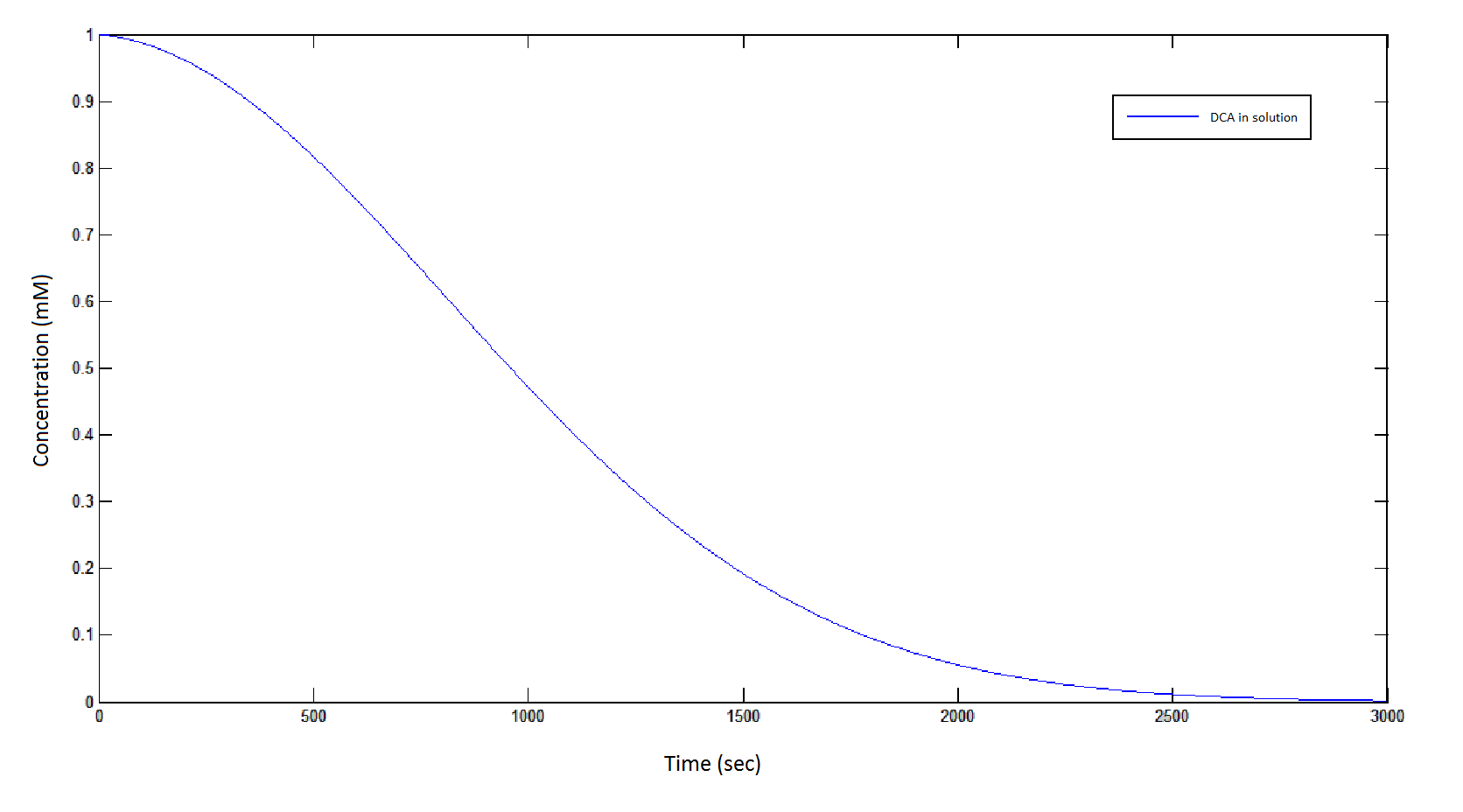
The rate at which DCA is removed in solution:
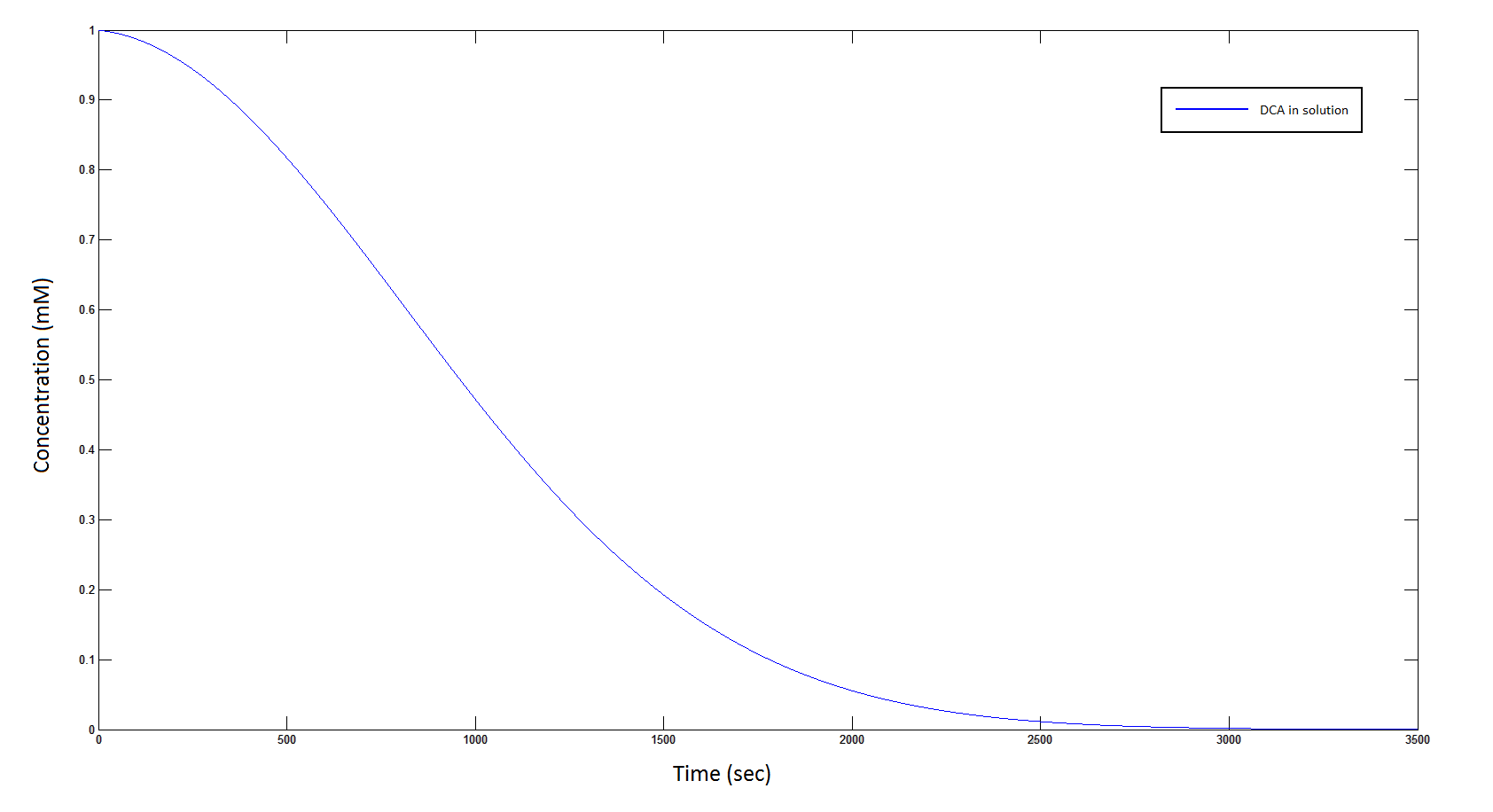
Graph 1: The blue line represents how the concentration of DCA in solution (extracellular) decreases due to the action of our DCA degraders. One can see that an initial concentration of 2E8 cells/mL completely removes the DCA with a concentration 1mM in (roughly) 150mins.
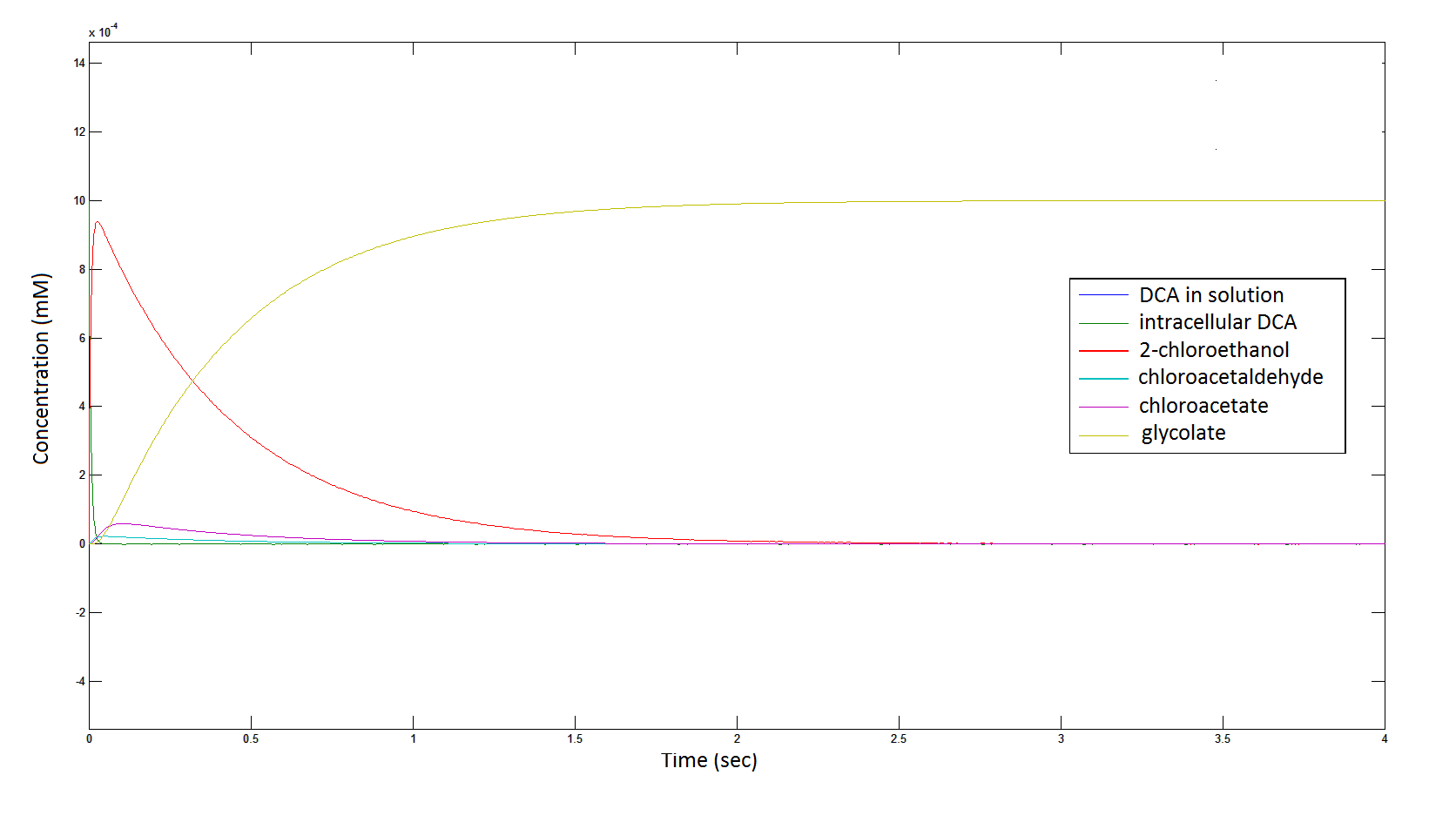
The Rate at which the intracellular concentration of the metabolites change over time:
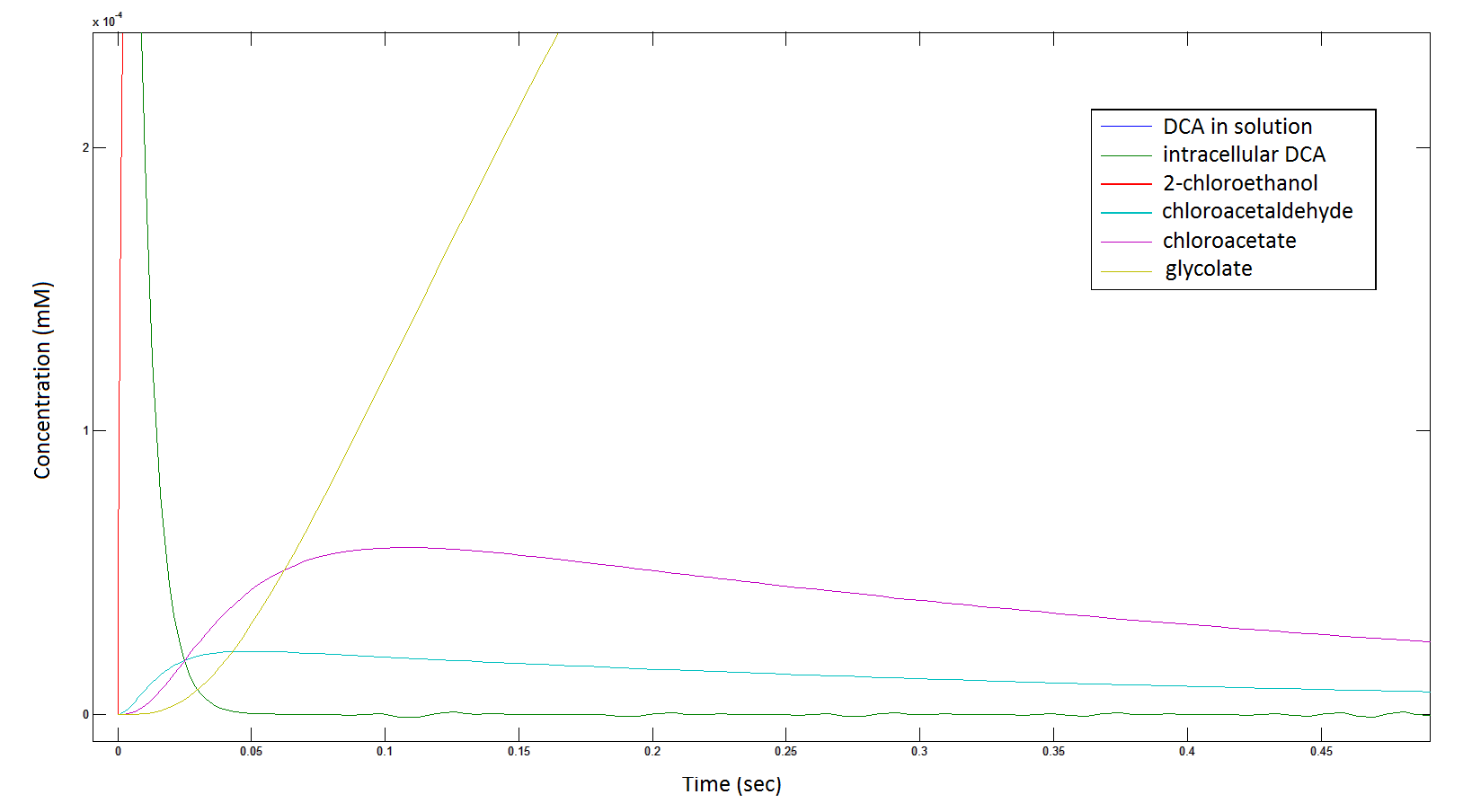
Graph 2: Each line represents the concentration of each of the metabolites. This graph is simply a rescaling of graph 1. Note: the glycolate won’t accumulate in the cell because it is metabolised. The model had glycolate as the final product and can be used to show how the presence of glycolate can lead to cell growth.
Rescaling the graph once again: the rate at which the metabolic intermediate change over time.
Graph 3: Each line represents the concentration of each of the metabolites . This graph is simply a rescaling of graph 1 and 2.
The rate at which the cells grow over time:
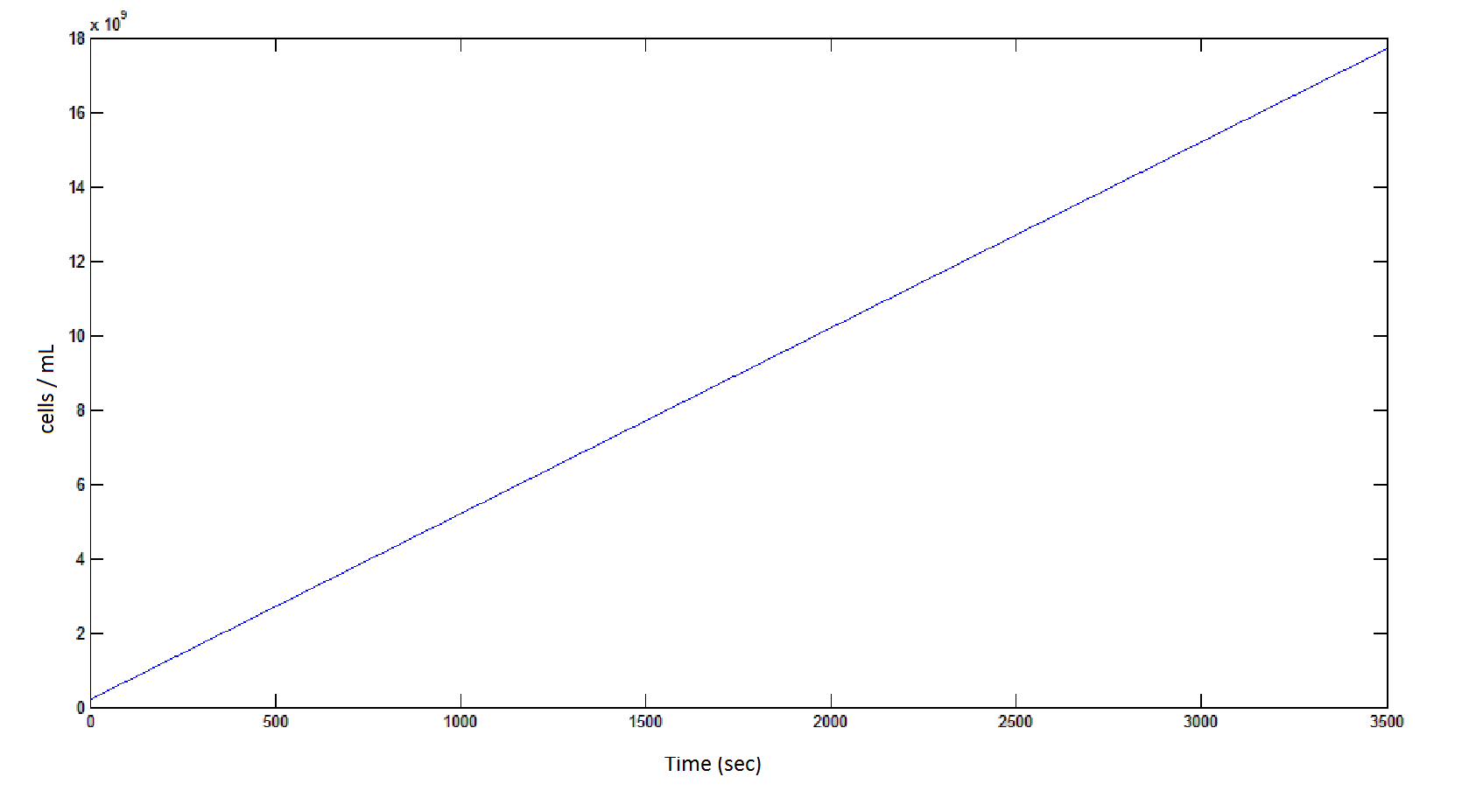
Graph 4: the blue line represents the linear increase of cells due to the presence of intracellular glycolate.
The Monooxygenase Pathway
ODE overview:
Again, this system of ODE has been explained and justified in the previous section. Below it is converted to MATLAB, allowing a computer to generate a visual representation of the model.
Raw MATLAB code:
function dy = p450(t,y)
dy=zeros(6,1);
dy(1)= -y(6)*(6*10^(-12))*0.0463067*(y(1)-y(2));
dy(2)= ((6*10^(-12))*0.0463067*(y(1)-y(2))) - 0.0113*25.55*(y(2)/(.12+y(2)));
dy(3) = 0.0113*25.55*(y(2)/(.12+y(2))) - 0.6*25.55*(y(3)/(0.16+y(3)));
dy(4)= 0.6*25.55*(y(3)/(0.16+y(3))) - 25.4*25.55*(y(4)/(20+y(4)));
if y(5) > 2*10^(-10)
dy(5)= 25.4*25.55*(y(4)/(20+y(4))) -1.5789*10^(-10)
else
dy(5) = 25.4*25.55*(y(4)/(20+y(4)))
end
if y(5) > 0.0005
if y(6) > 1.6*10^11
dy(6)=0
else
dy(6) = 5*10^6
end
else
dy(6) = 0
end
end
MATLAB output:
The rate at which DCA is removed in solution:
Graph 5: The blue line represents how the concentration of DCA in solution (extracellular) decreases due to the action of our DCA degraders.
The Rate at which the intracellular concentration of the metabolites change over time:
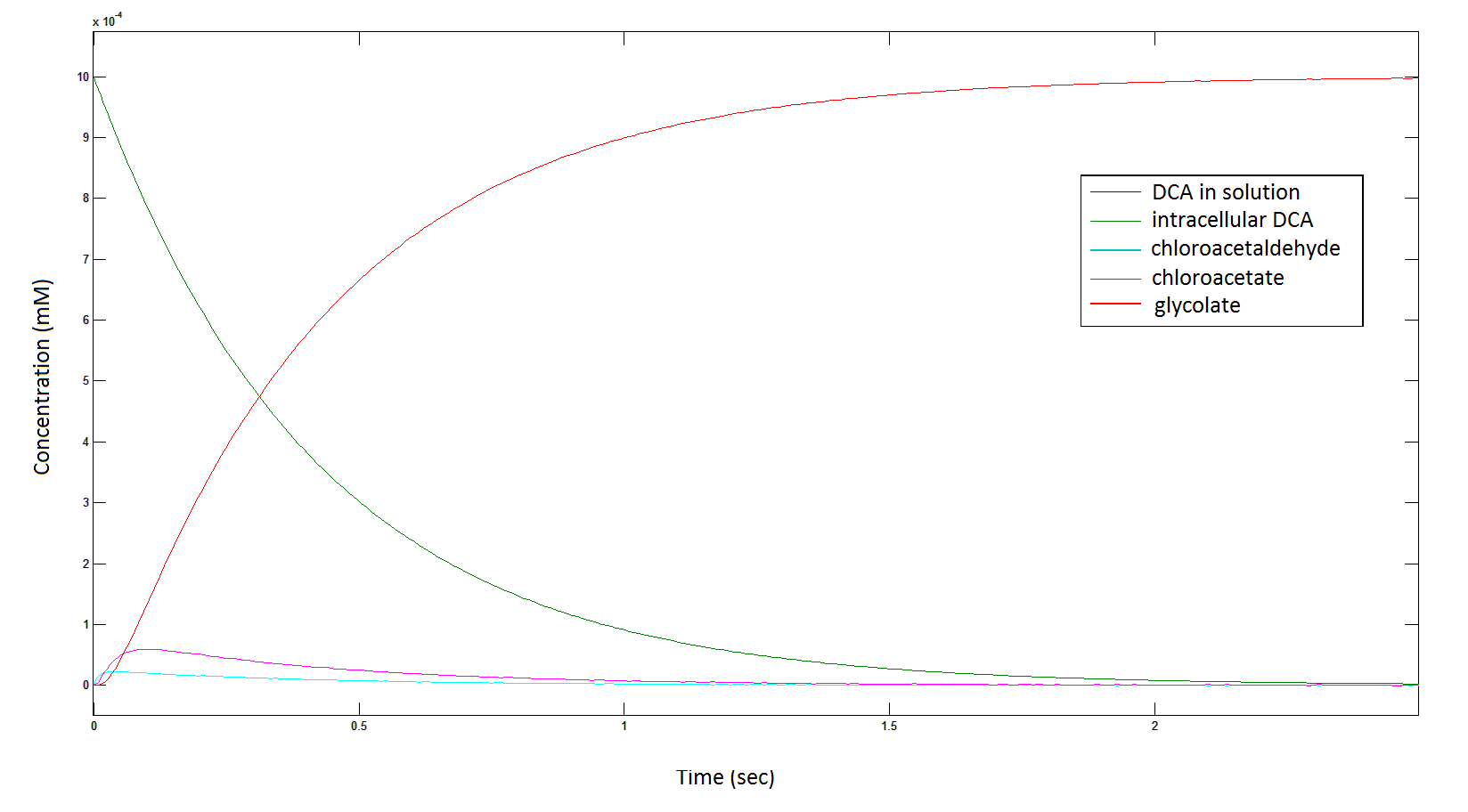
Graph 6: Each line represents the intracellular concentration of each of the metabolites over time within a single cell. The colour associated with each metabolite is depicted in the figure legend. This graph is simply a rescaling of graph 5.
Rescaling the graph once again: the rate at which the metabolic intermediate change over time.
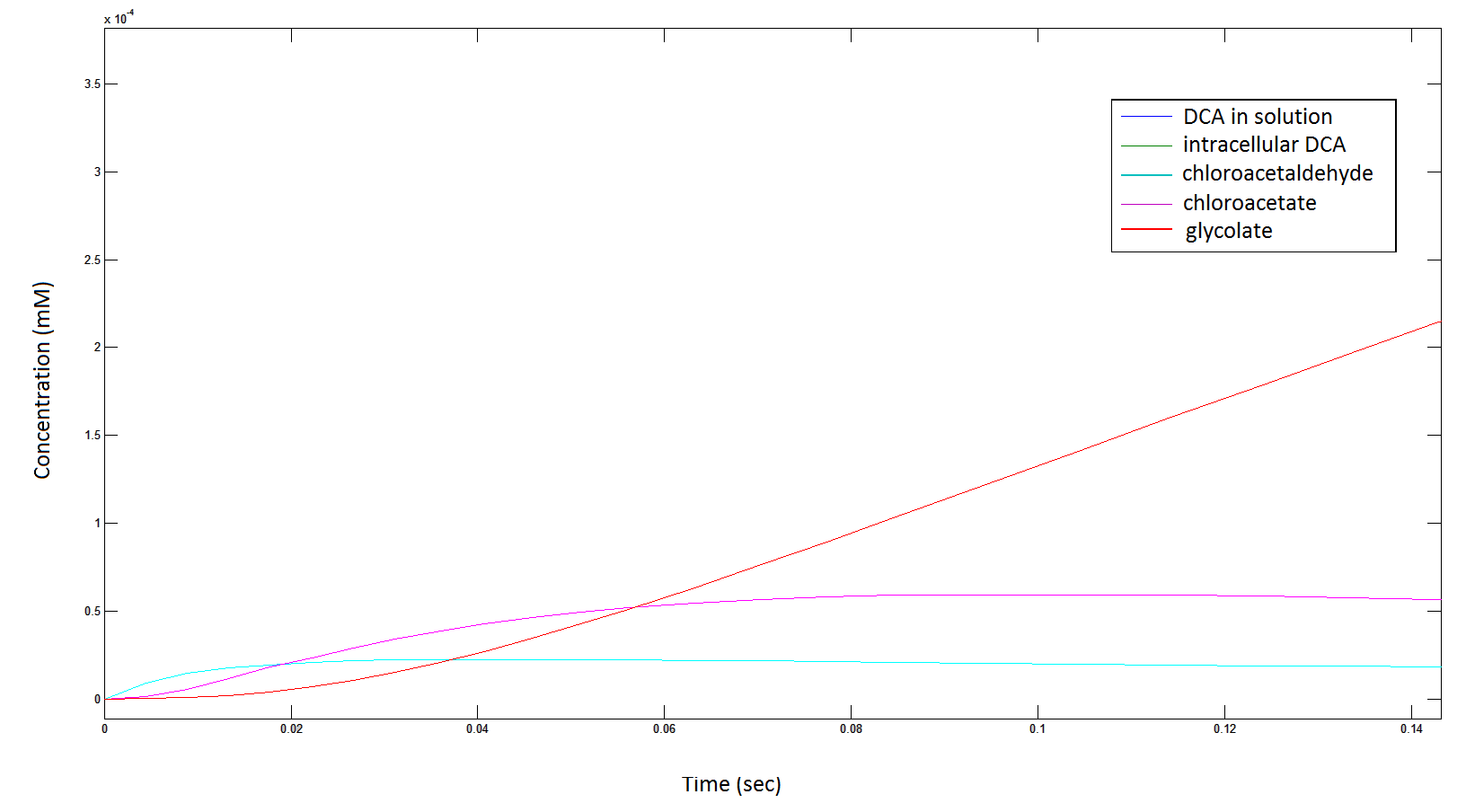
Graph 7: Again, each line represents the intracellular concentration of each of the metabolites over time within a single cell. This graph is simply a rescaling of graph 5 and 6.
The rate at which the cells grow over time:
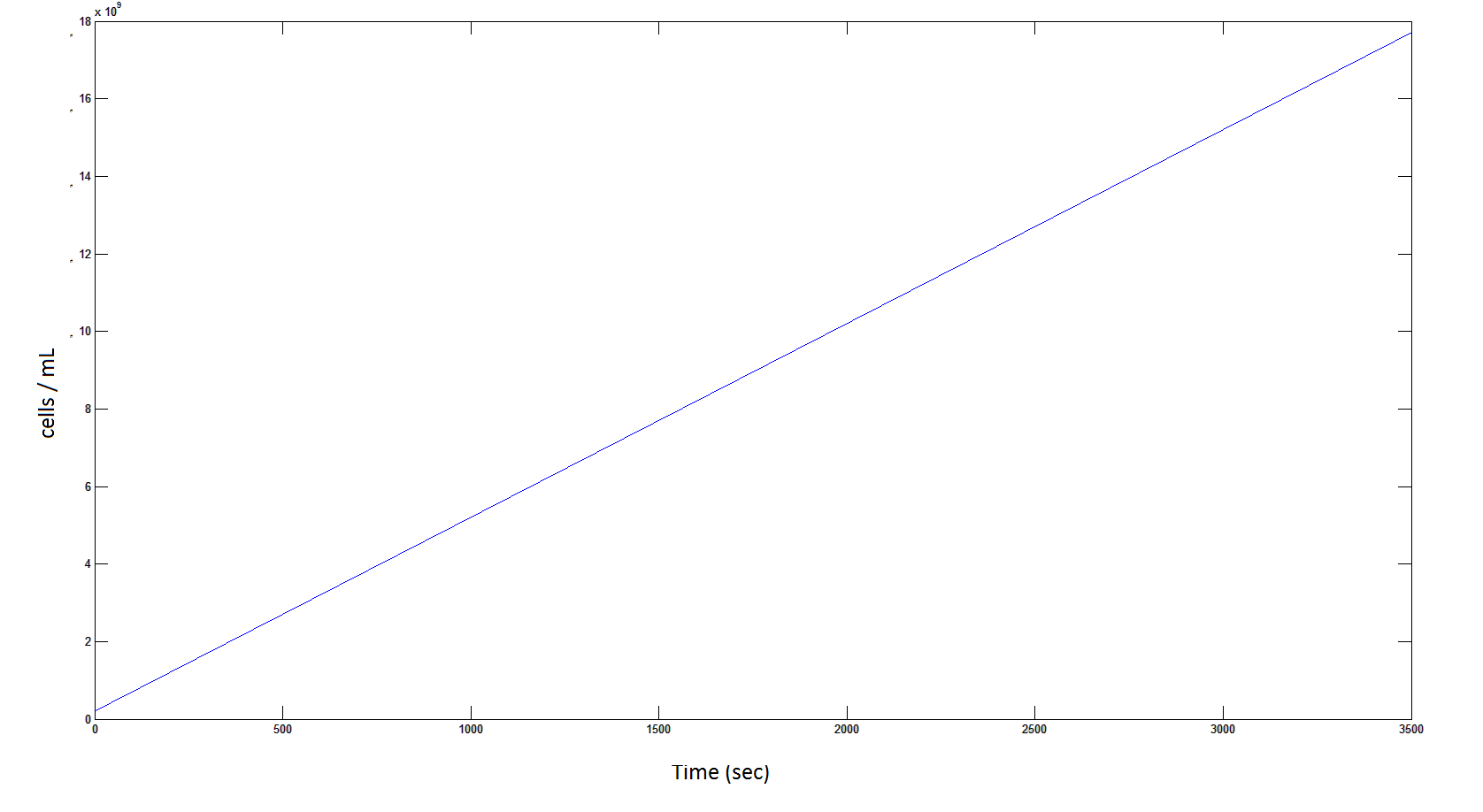
Graph 8: The blue line represents the linear increase of the total number of cells due to the presence of intracellular glycolate.
Rescaling Shows a lag effect on cell growth:
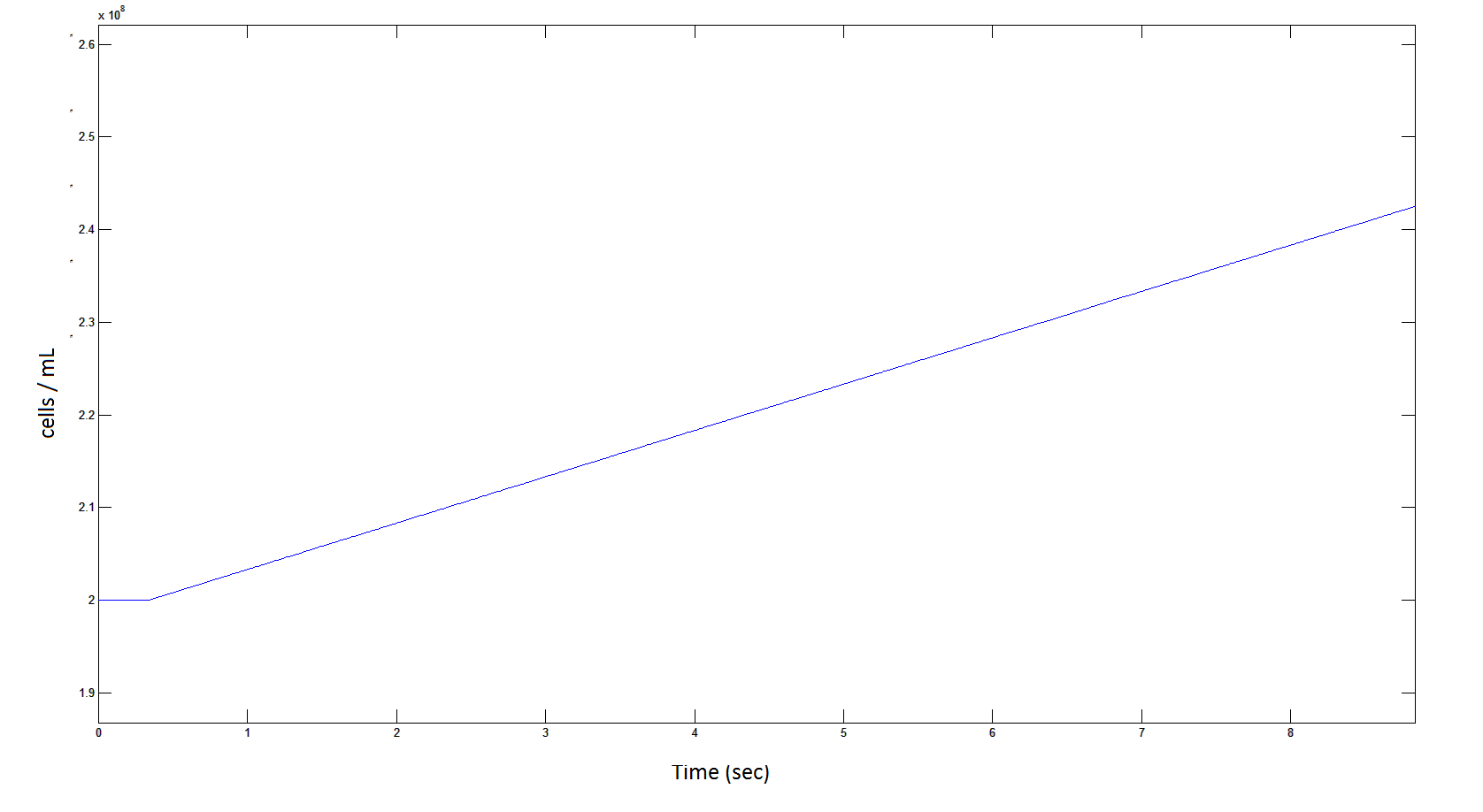
Graph 9: The blue line represents the increase of cells due to the presence of intracellular glycolate. This graph is a rescaling of graph 8 to show the inital lag in cellualr growth.
 "
"