Exeter/19 September 2013
From 2013.igem.org
OutPut Bricks formed using IDT gBlocks
The gBlocks for our output brick were ligating using the Gibson Assembly protocol. The output brick is responsive to red light, as it contains the specific promoter ([http://parts.igem.org/Part:BBa_R0082?title=Part:BBa_R0082 BBa_R0082]) for the Red Light Module (see our theory page).
The structure of the output brick is such; the specific promoter to correspond to the Red Light Module, a restriction site for the enzyme BglII, an RBS, the coding region for [http://parts.igem.org/wiki/index.php?title=Part:BBa_E0040 GFP], a restriction site for the enzyme BamHI and a series of STOP codons to act as a terminator. A simple diagram is given below.
(insert diagram)
The gBlocks were ligated and transformed on a previous day; this morning we looked at the liquid cultures we had made of them and noticed that, after centrifuging the cultures in preparation for Miniprepping, some of the cultures had a blue/green tinge. We exposed them to UV light; they were fluorescing green with varying shades of brightness. As the construct is formed from two gBlocks, with the cut between the two some way into the coding region for GFP, this shows that the gBlocks have correctly ligated as GFP is clearly being expressed.
This response, however, is not light-dependant. We should only be getting GFP when the cells are not exposed to red light, but as these cultures were grown in white light (which includes red light) and expressing GFP, this isn't happening. This is because we have transformed the gBlocks into regular competent cells (XL1 or DH5α) which produce the protein which switches on the promoter in front of GFP as part of their normal cellular functions (see our theory page for a more in-depth description of the Red Light Module).
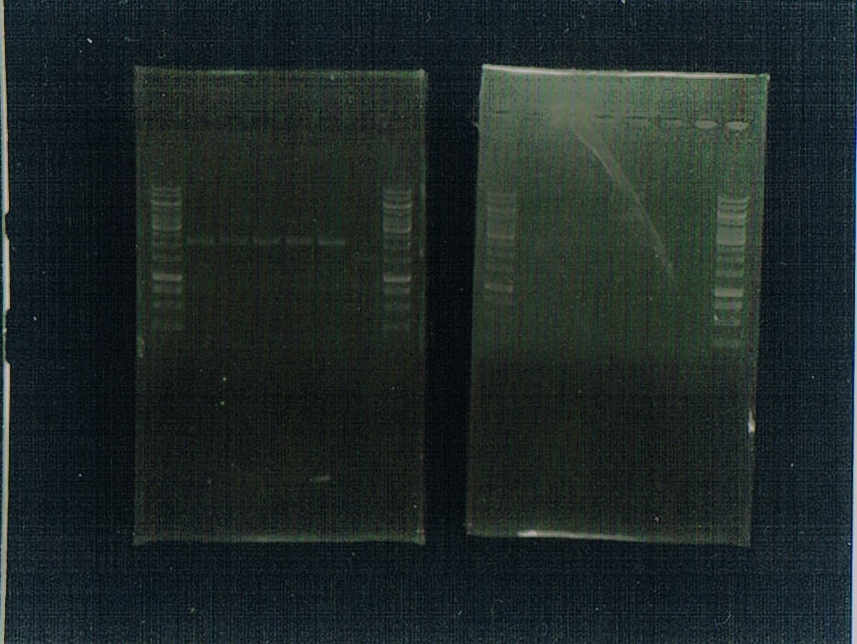
To confirm that we have functioning BglII and BamHI restriction sites within our gBlocks, we then digested 12 samples from our liquid cultures. They were Miniprepped and the cultures that gave the highest NanoDrop values (24A and 26B) were kept aside to be sent to the iGEM registry. The remaining 12 cultures were divided in two; 6 were digested EcoRI, PstI, BglII and BamHI (cultures 21A, 22A, 23A, 25A, 26A and 27A) and 6 with BglII and BamHI alone (cultures 21B, 22B, 23B, 24B, 25B and 27B).
Gel 1, cut with EcoRI, PstI, BglII and BamHI
| Lane | Contents |
|---|---|
| 1 | 1kb GeneRuler Ladder |
| 2 | 21A |
| 3 | 22A |
| 4 | 23A |
| 5 | 25A |
| 6 | 26A |
| 7 | 27A |
| 8 | 1kb GeneRuler Ladder |
Gel 2, cut with BglII and BamHI
| Lane | Contents |
|---|---|
| 1 | 1kb GeneRuler Ladder |
| 2 | 21B |
| 3 | 22B |
| 4 | 23B |
| 5 | 24B |
| 6 | 25B |
| 7 | 27B |
| 8 | 1kb GeneRuler Ladder |
Results of gels
Gel 1 had worked; there is a clear band for the CAM plasmid and faint (but present!) bands for the gBlocks. Gel 2 sadly didn't work, despite the ladders running. However, we are sure the gBlocks have ligated correctly as GFP is being produced; this just means we are uncertain about the BglII and BamHI restriction sites.
Take me back to the notebook.
 "
"