Team:Buenos Aires/ biological design
From 2013.igem.org
Our biological design
| Our aim was to get a final product easy to use and to understand, cheap and distributable to assess arsenic (and other pollutants) concentration in water, by anyone with no need of special training or other specific machines.
To determine the concentration of pollutants, our system would be based on the comparison of our sample with a known concentration curve. |
First issue: the output
The first choice we had to make was what kind of output would our system have. We wanted an output visible to the naked eye, so we had two main options: using pigment producing enzymes or coloured fluorescent proteins. One major difference between using enzymes for producing the output instead of coloured proteins is that the signal is amplified, as each enzyme unit is capable to produce many pigment molecules. This may also be a risk, as a high enzymatic activity can lead to saturation of the signal. On the other hand, pigment synthesis pathways are multi enzymatic so it requires several proteins to get to the final product. Consequently, it implies a need for more genes and more time to get the output signal.
Considering all these points, we finally decided to use the fluorescent protein mRFP that is also visible by the naked eye.
Second issue: signal production dependency to external conditions
The second problem we faced was that the protein production depends on several external conditions, as temperature, number of bacteria, etcetera. For instance, a same concentration of arsenite could lead to different colour intensities in different climate conditions.
To solve this problem, we needed the standard pattern of our physical device to be turned on at the same time as the sample in study. For these, our device would include dry wells with lyophilized bacteria and arsenite at different amounts, which should be resuspended with distilled water at the same time as the sample of water is added to the lyophilized bacteria in the well meant for the unknown sample.
As the whole device is subjected to the same conditions, if it is exposed to anything that causes cell death, as extreme heat, the effect would be homogeneous for all the wells.
Third issue: amplification of the signal and avoiding saturation
We performed some starting experiments to know if the protein mRFP under the arsenite inducible promoter pArs ([http://parts.igem.org/Part:BBa_K1106003 Bba_K1106003]) fulfilled our needs.
When testing fluorescence with a fluorimeter, this promoter showed an arsenite dose response sufficiently sensitive for our needs. Nevertheless, this differences where not appreciable by the naked eye. In addition, it took more than one day to get a clear red signal at high arsenite concentrations.
We identified the need of increasing the production rate of mRFP, but this first design is not flexible according to mathematical analysis. But the problem with amplification is that it may lead to quick saturation of the output signal.
We discussed some alternative designs and finally got to a design that:
• Includes a step of signal amplification
• Stops output production at a certain moment before saturation.
| It is important to consider that if our input had been an enzymatically synthesized pigment, stopping the protein production wouldn’t have been enough to stop the output production, as enzyme molecules remaining could keep on synthesizing the pigment.
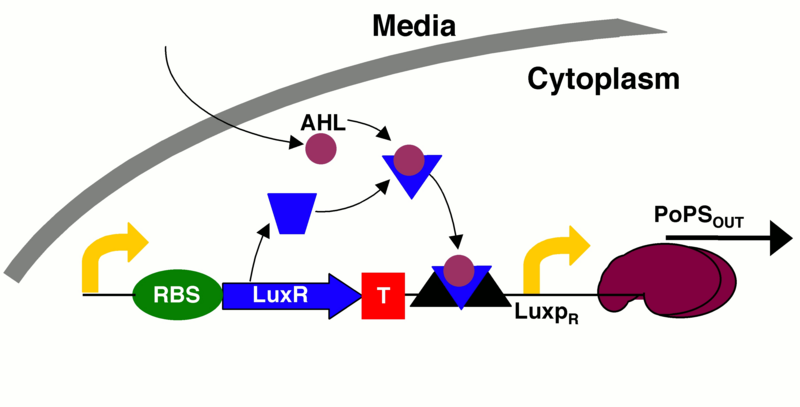
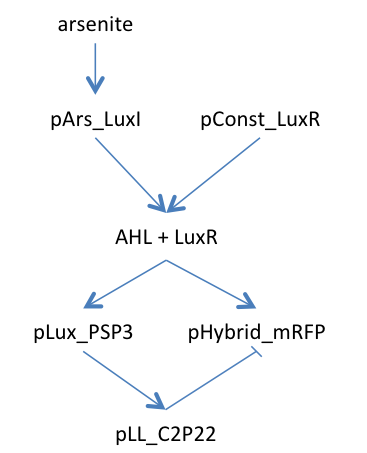
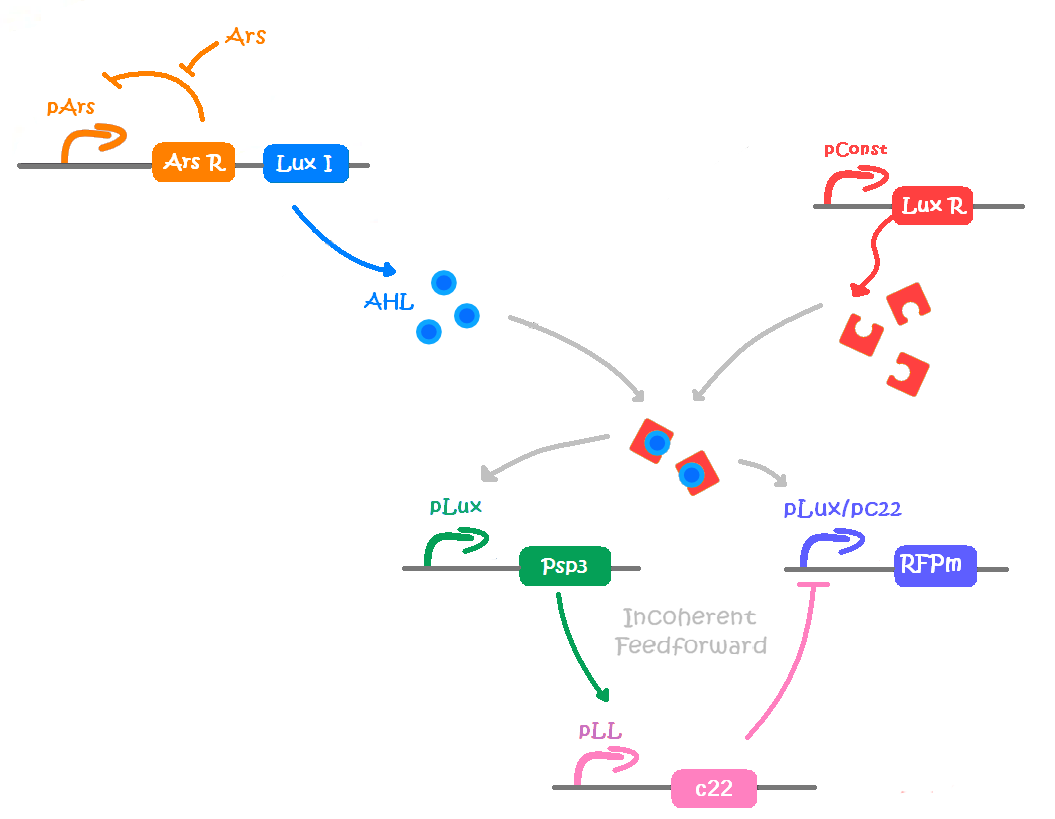
An incoherent feedforward scheme was chosen; it would satisfy our needs, as we can amplify the signal produced by the input, in our case the pollutant, and stop the production of the output signal through a delayed inhibition of the output production. Briefly, the input signal A induces the production of B and the output C, which, in turn, is inhibited by B with a time delay. In biological terms, if we want to start the production of a protein (the output) and then stop it, we need an inducible/repressible promoter. Among the options available in the Parts Registry, we found an hybrid promoter that would fit our requirements. It is activated by the [http://parts.igem.org/Lux Lux quorum sensing system] of Vibrio fischeri, and is repressed by the P2 phage transcription repressor [http://parts.igem.org/wiki/index.php?title=Part:BBa_K145150 P2C22].
|
LuxI would work as an amplifier, as each enzyme molecule can synthesize several AHL molecules.
As we need a delay to allow colour production in our incoherent feedforward system, we decided to introduce an intermediary protein to produce the repression. Thus, the expression of the repressor C2P22 under the regulation of the pLL promoter is induced by the transcription activator PSP3, which is under the regulation of the pLux promoter. According to the mathematical model results, this design is feasible.
The figure below synthetizes our model in a more graphic way:
In addition, as AHL is diffusible, the arsenite detection can be uncoupled from the output production in two different bacteria.
 "
"