Team:Heidelberg/Templates/Indigoidine week15
From 2013.igem.org
Contents |
CPEC Conditions Assay (Ralf)
We performed CPEC assembly of pRB13 with eight different CPEC conditions to see whether we get difeerent results.
Table 11.1 CPEC mix for assembly of pRB13
| Annealing Temp [°C] | Cycles | ||
|---|---|---|---|
| 5 | 10 | 15 | |
| 53 | 2: 10 ul; 4 ul backbone; 1 ul CCDB | ||
| 55 | 3: 10 ul; 4 ul backbone; 1 ul CCDB 7: 20 ul; 8.5 ul backbone; 1.5 ul CCDB | 4: 10 ul; 4 ul backbone; 1 ul CCDB 6: 20 ul; 8.5 ul backbone; 1.5 ul CCDB 8: 10 ul; 4.5 ul backbone; 0.5 ul CCDB | 5: 10 ul; 4 ul backbone; 1 ul CCDB |
| 57 | 1: 10 ul; 4 ul backbone; 1 ul CCDB | ||
Table 11.2 CPEC Assembly of pRB13
| BioRad T100 | ||
|---|---|---|
| cycles | temp [°C] | time [s] |
| 1 | 98 | 10 |
| 5/10/15 | 98 | 1 |
| 53/55/57 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
100 ul of competent TOP10 were transformed with 5 ul of reaction product and incubated for 30 hours at 37 °C.
Apparently there is no big difference using various CPEC conditions.
We will continue with the standard protocol.
PPTase test (Ralf)
pRB3-10 are plasmids containing indC (pRB-3-6) or bpsA(pMM64) (pRB7-10) and a PPTase (sfp, svp, svp(pMM65), entD).
This week the blue colonies are screened and sequenced for validation.
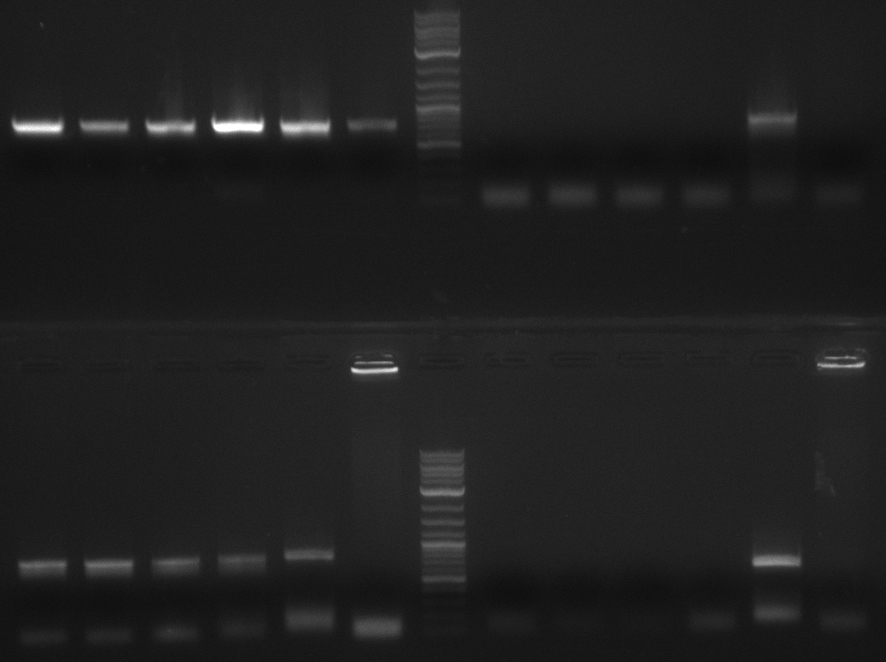
colony PCR screening #1
Two blue colonies are picked from each plate and screened with the primers of the PPTase they should have and the
ones which they could have if something went wrong. PPTase PCR products werde used as a positive control and TOP10
without any plasmid as a negative control. So numbers 5 to 18 are subdivided into alpha and beta.
| PPTase to be screened | ||||
|---|---|---|---|---|
| template | sfp (RB35/36) | svp (RB29/30) | svp(pMM65) (RB25/26) | entD (RB33/34) |
| 1 | ||||
| 2 | ||||
| 3 | ||||
| 4 | ||||
| 5 | ||||
| 6 | ||||
| 7 | ||||
| 13 | 14 | 15 | 8 | |
| 9 | ||||
| 10 | ||||
| 11 | ||||
| 16 | 17 | 18 | 12 | |
| 19 | 20 | 21 | 22 | |
| BioRad T100 | ||
|---|---|---|
| cycles | temp [°C] | time [s] |
| 1 | 120 | |
| 30 | 95 | 60 |
| 65 | 30 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
| Loading Scheme | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5a | 5b | 9a | 9b | 1 | 19 | 2-log | 6a | 6b | 10a | 10b | 2 | 20 |
| 7a | 7b | 11a | 11b | 3 | 21 | 2-log | 8a | 8b | 12a | 12b | 4 | 22 |
The gel is not conclusive. Therefore I will repeat the screening but screen every colony for all PPTases.
MiniPrep of pRB4-10
5 ml liquid cultures of TOP10+pRB4-10 have been miniprepped using QIAquick miniprep kit. Only blue cultures have
been prepped (except pRB9 because there was no blue culture). pRB4-6 and pRB10 were prepped in duplicates (liquid
cultures of two different clones) whereas there are only single preps of pRB7-9. This is because pRB3-6 contain
indc ahich we will use for further studies. pRB6 and 10 contain entD, which is the E. coli endogenous PPTase and
was previously declared to be "inefficient" in activating bpsA [Takahashi 2007].
Concentrations of the prepped plasmids have been measured using ??ThermoScientific NanoDrop
| plasmid | version | concentration [ng/ ul] |
|---|---|---|
| pRB4 | alpha | 289.7 |
| beta | 278.7 | |
| pRB5 | alpha | 228.3 |
| beta | 348.9 | |
| pRB6 | alpha | 428.0 |
| beta | 432.2 | |
| pRB7 | alpha | 187.3 |
| pRB8 | alpha | 768.2 |
| pRB9 | alpha | 142.5 |
| pRB10 | alpha | 112.0 |
| beta | 212.0 | |
| water after measurement | 0.7 | |
Minipreps were reduced to a concentration of about 50 ng/ ul for sequencing and 1 ng/ ul for further studies.
MiniPrep PCR screening #2
| PPTase to be screened | ||||
|---|---|---|---|---|
| template | sfp (RB35/36) | svp (RB29/30) | svp(pMM65) (RB25/26) | entD (RB33/34) |
| TOP10+pRB4 | 5 | 6 | 7 | 8 |
| TOP10+pRB5 | 9 | 10 | 11 | 12 |
| TOP10+pRB6 | 13 | 14 | 15 | 16 |
| TOP10+pRB7 | 17 | 18 | 19 | 20 |
| TOP10+pRB8 | 21 | 22 | 23 | 24 |
| TOP10+pRB9 | 25 | 26 | 27 | 28 |
| TOP10+pRB10 | 29 | 30 | 31 | 32 |
| sfp | 33 | |||
| svp | 34 | |||
| svp(pMM65) | 35 | |||
| entD | 36 | |||
| TOP10 | 37 | 38 | 39 | 40 |
| BioRad T100 | ||
|---|---|---|
| cycles | temp [°C] | time [s] |
| 1 | 95 | 120 |
| 30 | 95 | 60 |
| 60 | 30 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
| Loading Scheme | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5a | 5b | 9a | 9b | 13a | 13b | 17 | 21 | 25 | 29a | 29b | sfp | 2-log | 6a | 6b | 10a | 10b | 14a | 14b | 18 | 22 | 26 | 30a | 30b | svp |
| 7a | 7b | 11a | 11b | 15a | 15b | 19 | 23 | 27 | 31a | 31b | svp(pMM65) | 2-log | 8a | 8b | 12a | 12b | 16a | 16b | 20 | 24 | 28 | 32a | 32b | entD |
pRB9 is negative. All screens are positive in the svp(pMM65) and entD section. In entD this could be due to genomic
DNA in the miniprep. svp(pMM65) is also positive in all plasmids, which is strange. Maybe the PCR conditions have
been too tolerant. Nevertheless the bands of the plasmids which should carry the respective PPTase appear to be
stronger. We will sequence the plasmids anyways.
Sequencing of pRB3-10
plasmid minipreps have been sequenced by GATC [reference] using the sequencing primers VF2 and VR. The sequencing
results show that all the inserts are correct except for pRB9, which isn't a pity because we have previously shown
with pKH1-2 that the combination of bpsA(pMM64) and svp(pMM65) successful produces indigoidine.
| plasmid | Primer | ind synthetase/ PPTase | sequencing result/ reference |
|---|---|---|---|
| pRB3 | VF2 | indC | |
| seq2 | |||
| VR | sfp-rev | ||
| seq2 | |||
| pRB4 | VF2 | indC | |
| seq2 | |||
| VR | svp(pMM65)-rev | ||
| seq2 | |||
| pRB5 | VF2 | indC | |
| seq2 | |||
| VR | svp-rev | ||
| seq2 | |||
| pRB6 | VF2 | indC | |
| seq2 | |||
| VR | entD-rev | ||
| seq2 | |||
| pRB7 | VF2 | bpsA | |
| seq2 | |||
| VR | sfp-rev | ||
| seq2 | |||
| pRB8 | VF2 | bpsA | |
| seq2 | |||
| VR | svp(pMM65)-rev | ||
| seq2 | |||
| pRB9 | VF2 | bpsA | |
| seq2 | |||
| VR | svp-rev | ||
| seq2 | |||
| pRB10 | VF2 | bpsA | |
| seq2 | |||
| VR | entD-rev | ||
| seq2 |
In the following pRB4-beta, pRB5-alpha, pRB5-beta, pRB6-alpha and pRB10-alpha have been selected so that there are
single versions of pRB3-10 for furhter experiments.
Glycerols stocks pRB3-10
3 stocks each were made using 2 mL liquid cultures (48 h at 37 °C 200 rpm) of TOP10+pRB3-10, respectively.
 "
"