Team:Heidelberg/Templates/Indigoidine week16
From 2013.igem.org
Contents |
Preparation for T-Domain exchanges
We assemble pRB14, which is a version of pRB3, in which the T-Domain is exchanged by the whole insert of the pDONR vector. Since pRB13 grew on DH5alpha and TOP10 we hope that pRB14 won't. This vector will be used to exchange T-Domains and will be modified to get a version without the PPTase sfp and the cutting sites EcoRI and SpeI in indC. To check whether a successfully exchanged T-Domain can be activated by a defined PPTase, we build pSB2K3-derived plasmids with a single PPTase (sfp, svp, entD or delC) under control of the lac-Promotor and the RBS BBa_B0029.
Fragment Amplification
PCR of Fragments for PPTase plasmids
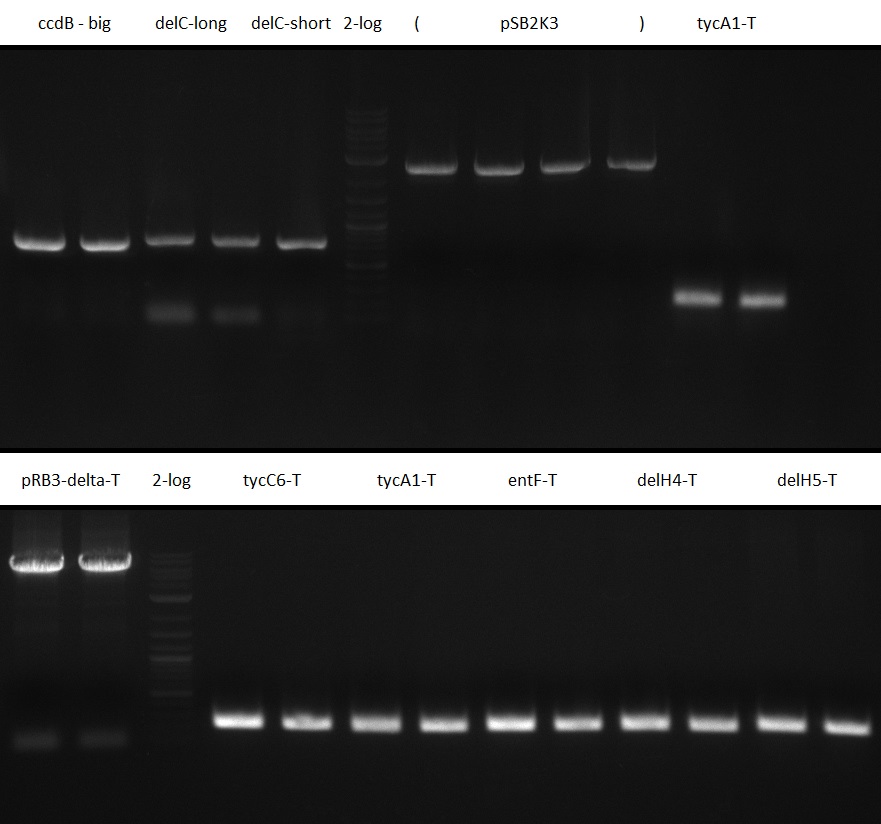
Table 12.x PCR for pRB15-18: 25 ul Phusion Flash HF MM 2x; 5 ul Primer 10 uM each; template according to table; water ad 50 ul.
| pSB2K3 RB21/63 in BioRAD old old | ||
|---|---|---|
| template: 3 ul pSB2K3 250 ng/ ul (distribution) | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 62 ? 0.5 | 5 | |
| 72 | 40 | |
| 25 | 98 | 1 |
| 57 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
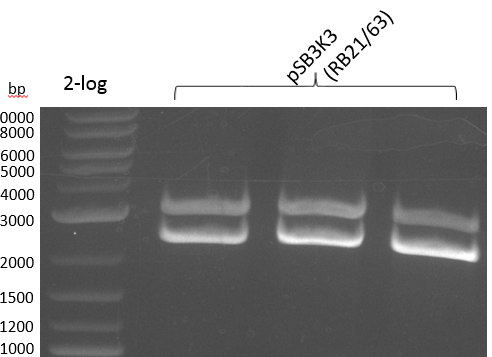
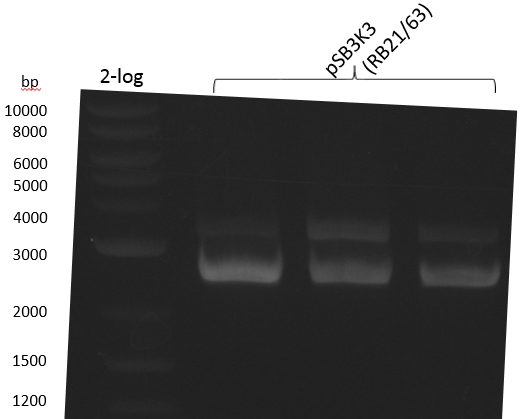
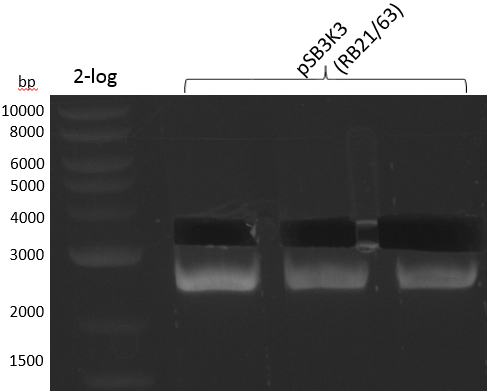
pSB2K3 contains the 3'-part of the reversed Primer in an internal lac-Operator-structure, so the PCR product is just half of the backbone (2500 bp instead of 4800 bp). We will use the same primers but pSB3K3 of the 2013 spring distribution plate 5 well 5E; which is around 3000 bp.
Table 12.x PCR for pRB15-18: 25 ul Phusion Flash HF MM 2x; 5 ul Primer 10 uM each; template according to table; water ad 50 ul.
| pSB3K3 RB21/63 in BioRAD old old | ||
|---|---|---|
| template: 2 ul pSB3K3 250 ng/ ul (distribution) | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 3 | 98 | 1 |
| 58 | 5 | |
| 72 | 50 | |
| 32 | 98 | 1 |
| 65 | 5 | |
| 72 | 50 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
We get unspecific product -> repetition with more stringent conditions
| pSB3K3 RB21/63 in BioRAD T100 | ||
|---|---|---|
| template: 0.5/1.5 ul pSB3K3 250 ng/ ul (distribution) | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 65 -0.5 | 5 | |
| 72 | 50 | |
| 25 | 98 | 1 |
| 69 | 5 | |
| 72 | 50 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
PCR of Fragments for T-Domain exchange
Table 12.x PCR for pRB13: 25 ul Phusion Flash HF MM 2x; 5 ul Primer 10 uM each; template according to table; water ad 50 ul.
| pRB3?T KH3/4 in T100 | ||
|---|---|---|
| template: 1 ul pRB3 1 ng/ ul | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 62 ? 0.5 | 5 | |
| 72 | 120 | |
| 25 | 98 | 1 |
| 57 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
| KH9/10 in T100 | ||
| template: 2 ul pDONR 1 ng/ ul | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 10 |
| 8 | 98 | 1 |
| 70 ? 0.5 | 5 | |
| 72 | 15 | |
| 28 | 98 | 1 |
| 65 | 5 | |
| 72 | 15 | |
| 1 | 72 | 90 |
| 1 | 12 | - |
| T-Domains in BioRAD old | ||
|---|---|---|
| tycC6-T RB57/58 1 ul B. para overnight culture | ||
| tycA1-T RB55/56 1 ul B. para overnight culture | ||
| entF-T RB53/54 colony MG1655 | ||
| delH4-T RB59/60 colony D. aci SPH-1 | ||
| delH5-T RB61/62 colony D. aci SPH-1 | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 120 |
| 40 | 98 | 1 |
| 65 | 5 | |
| 72 | 5 | |
| 1 | 72 | 30 |
| 1 | 12 | - |
Gel Extraction
Gel extraction was performed using QIAquick gel extraction kit. DNA yield was measured using NanoDrop Spectrophotometer ND-1000
Table 12.x DNA concentrations for assembly of pRB-T
| Fragment | Concentration [ng/ ul] | ~ Fragment size [bp] | Molarity [nM] |
|---|---|---|---|
| sfp | 329.6 | 700 | 713.42 |
| svp | 44.9 | 750 | 90.7 |
| entD | 289.2 | 650 | 674.13 |
| delC | 54.5 | 700 | 117.97 |
| pSB3K3 | 26.8 | 3000 | 13.54 |
| pRB3dT | 124.8 | 6800 | 27.81 |
| ccdB | 121.2 | 700 | 262.33 |
| indC-T | 192.4 | 200 | 1457.58 |
| bpsA-T | 198.9 | 200 | 1506.18 |
| entF-T | 184.8 | 200 | 1400.00 |
| tycA1-T | 184.2 | 200 | 1395.45 |
| tycC6-T | 198.6 | 200 | 1504.55 |
| delH4-T | 175.0 | 200 | 1325.76 |
| delH5-T | 162.4 | 200 | 1230.30 |
CPEC Assembly and Transformation
pRB14-18
| Plasmid | Fragment 1 | Molarity [nM] | Volume in MM | Fragment 2 | Molarity [nM] | Volume in MM |
|---|---|---|---|---|---|---|
| pRB14 | pRB3dT | 27.81 | 3.8 | ccdB | 262.33 | 1.2 |
| pRB15 | pSB3K3 | 13.54 | 4.5 | sfp | 713.42 | 0.5 |
| pRB16 | pSB3K3 | 13.54 | 3.5 | svp | 90.7 | 1.5 |
| pRB17 | pSB3K3 | 13.54 | 4.5 | entD | 674.13 | 0.5 |
| pRB18 | pSB3K3 | 13.54 | 3.8 | delC | 117.97 | 1.2 |
| pRB21 | indC(RB27/46) | 25.40 | 3.5 | pSB1C3(RB21/22) | 133.08 | 1.5 |
Table 12.x
- CPEC Assembly pRB14-18
| BioRAD T100 | ||
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 53 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
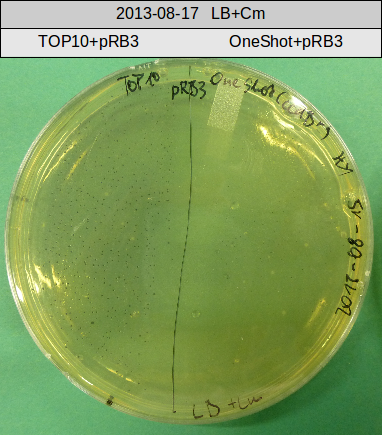
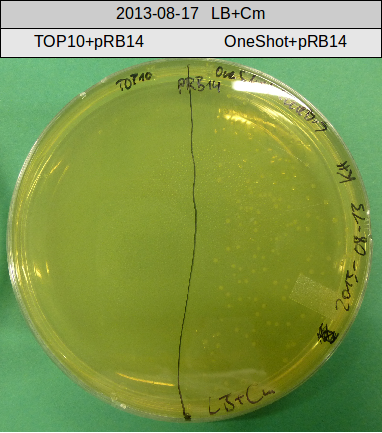
Transforation according to standard protocol for chemical transformation
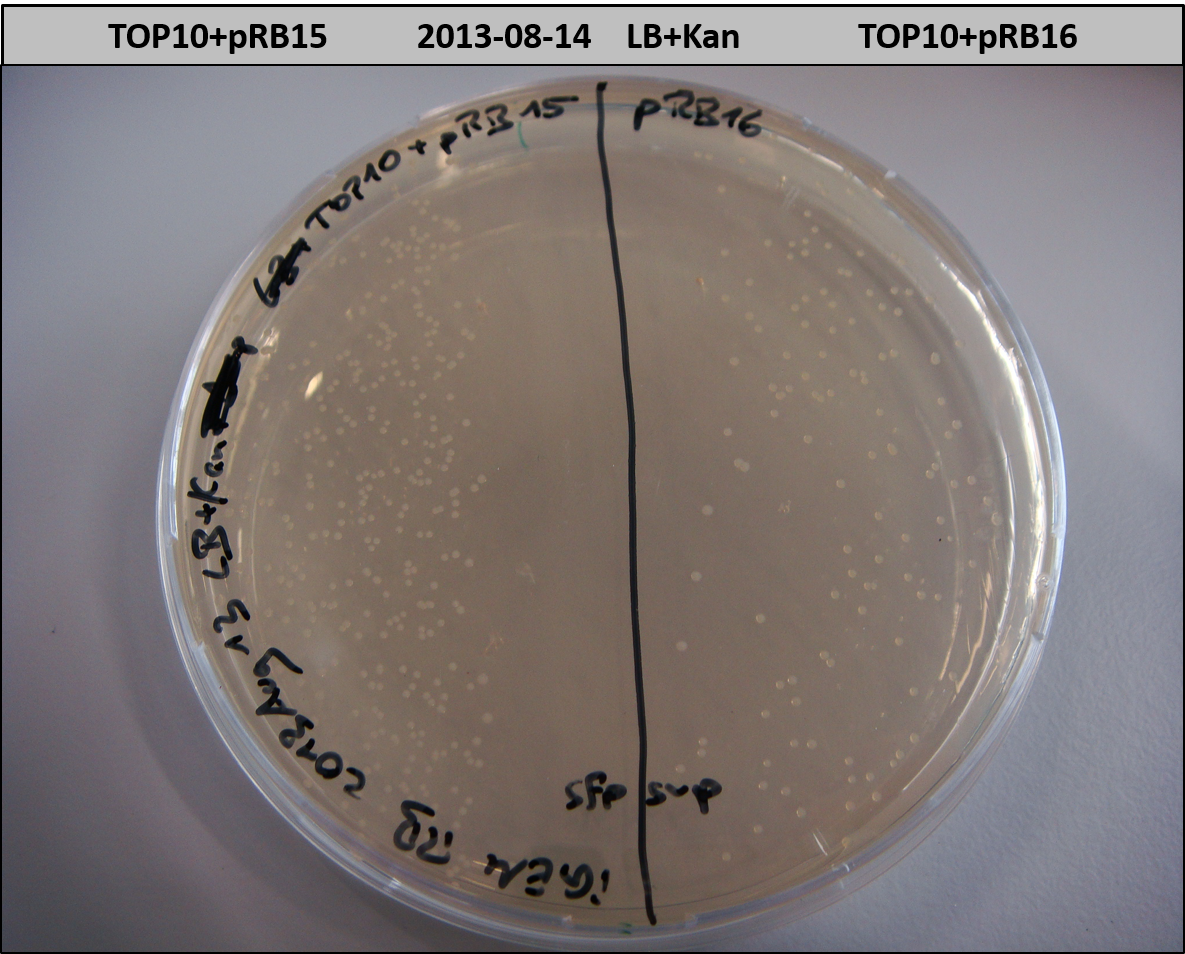
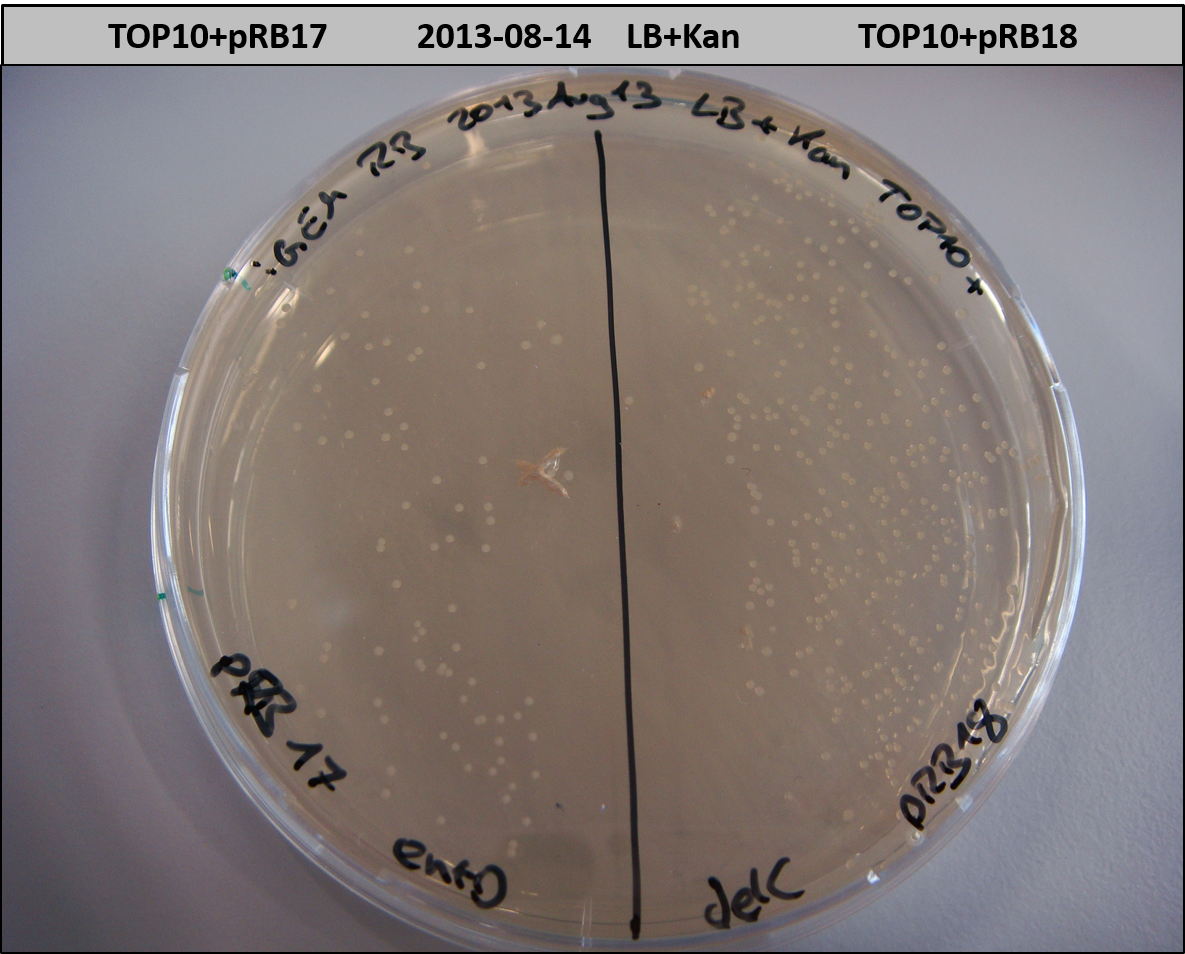
- TOP10 with pRB15-18 (-> LB+Kan)
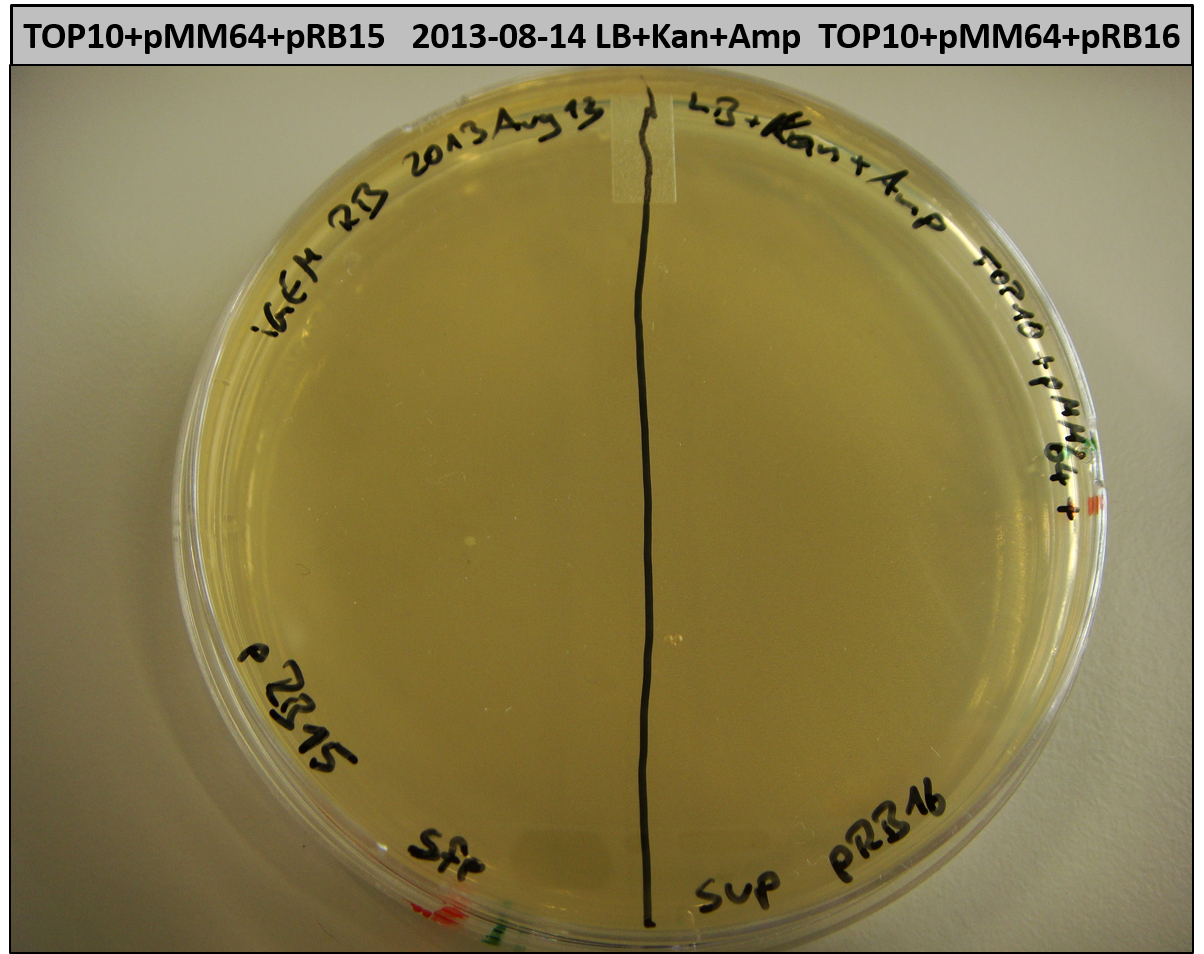
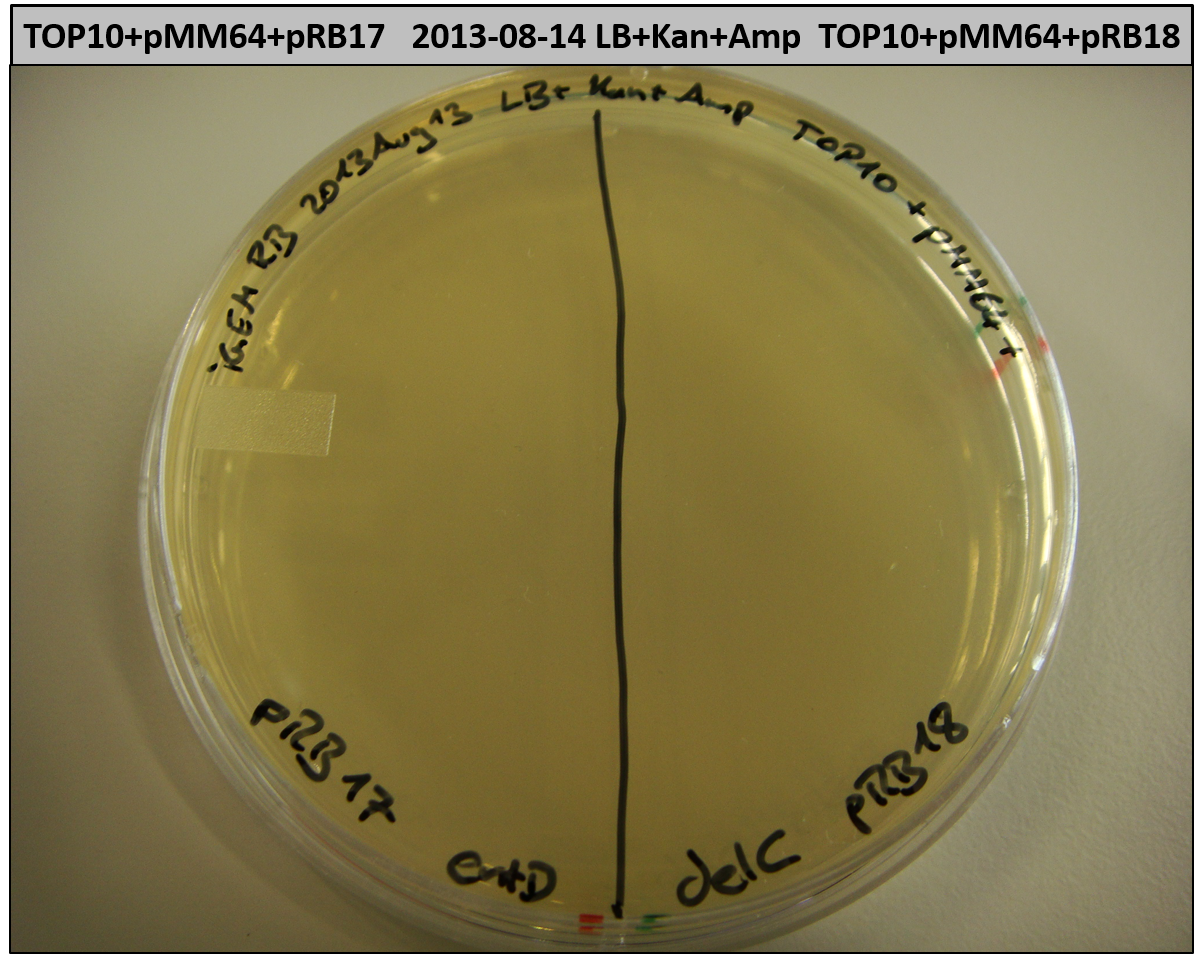
- TOP10 with pRB15-18 and pMM64, respectively (-> LB+Kan+Amp)
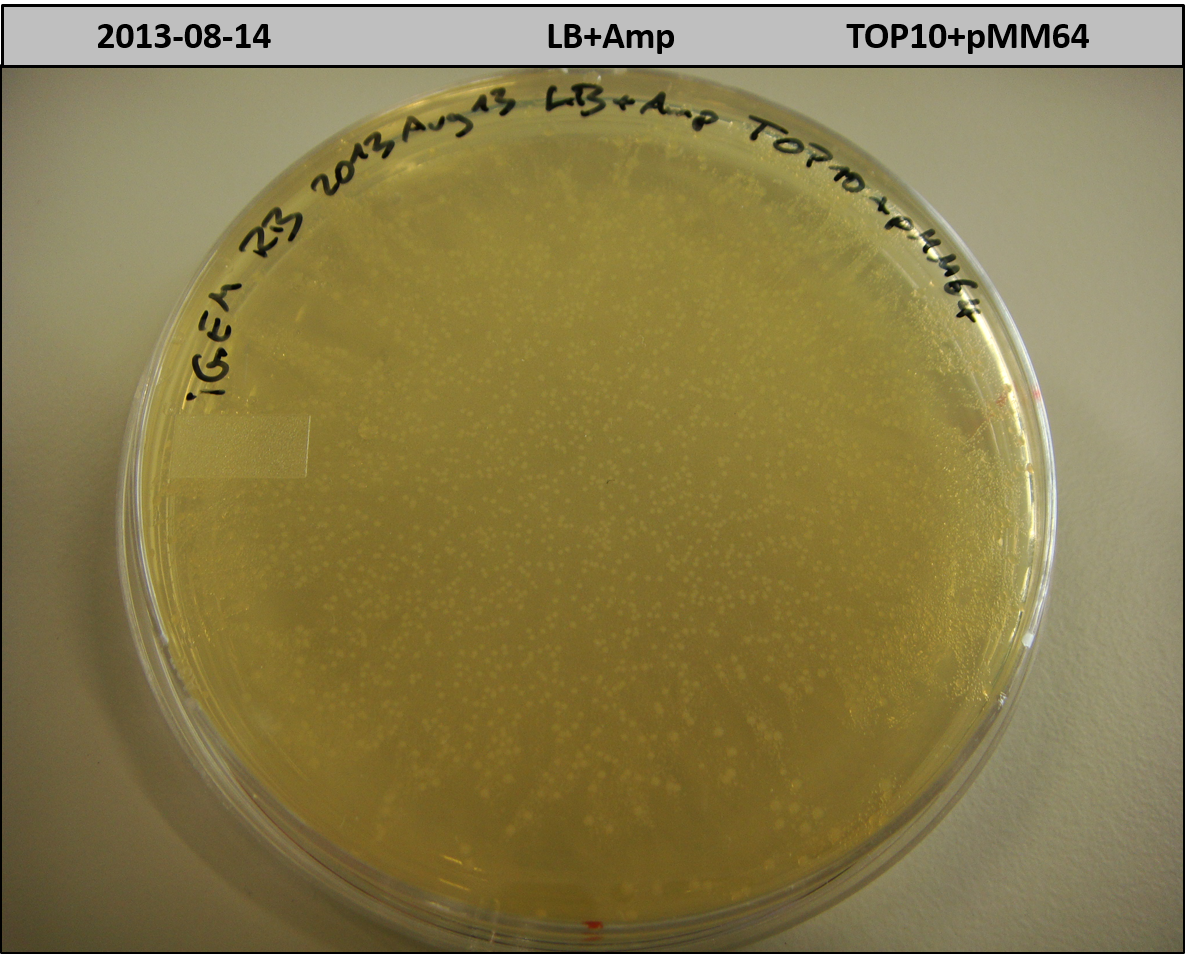
- TOP10 with pMM64 (-> LB+Amp)
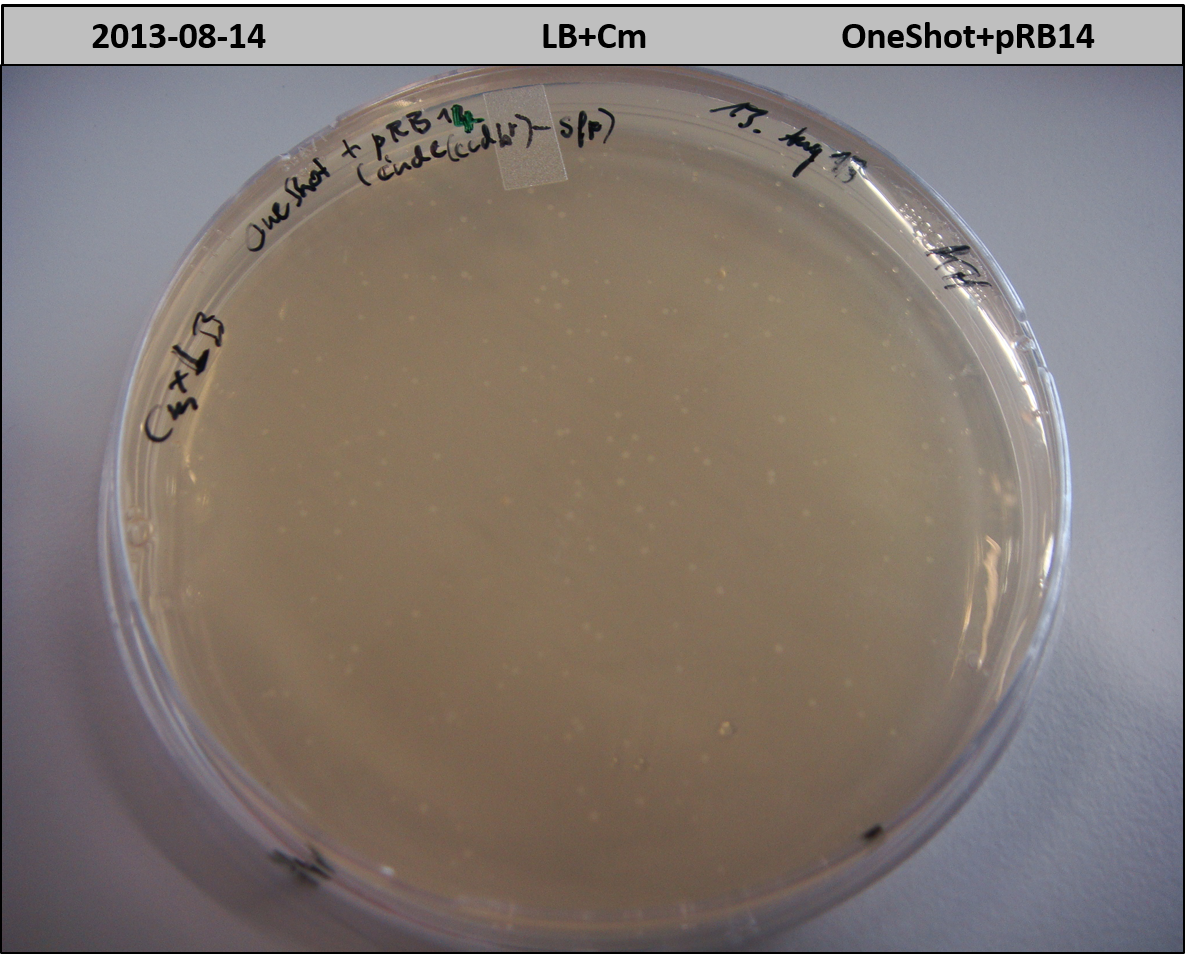
- OneShot with pRB14 (-> LB+Cm)
- OneShot+pRB14: small colonies after 20 hours
- TOP10+pRB15-18: colonies on all plates
- TOP10+pMM64+pRB15-18: (almost) no colonies after 20 hours; hopefully due to indigoidine growth retardation
- TOP10+pMM64 TraFo was efficient
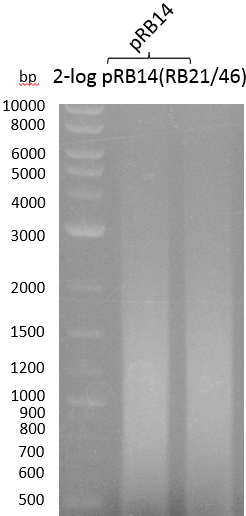
We will perform PCR screening in triplicates of TOP10+pRB15-19 and OneShot+pRB14 to get a first impression on whether the assembly was successful.
Plasmid Validation
PCR Screening
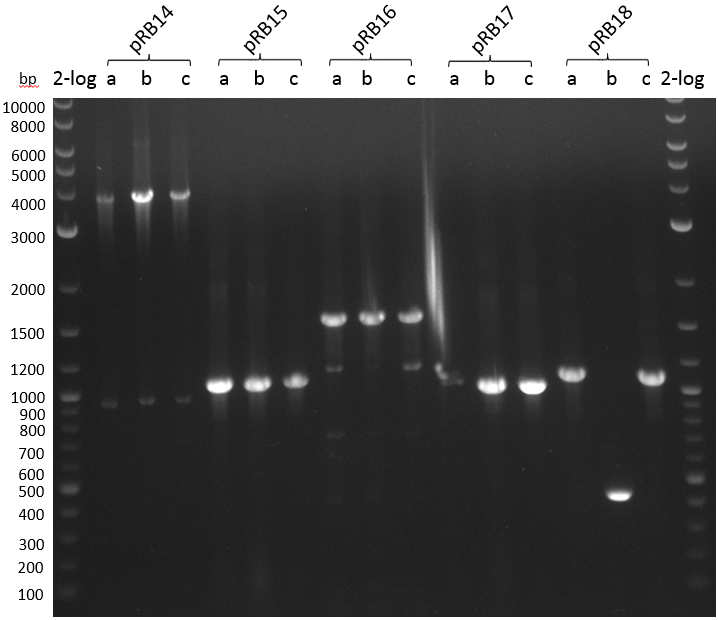
We use forward primers of the insert and standard reversed primer VR to screen pRB14-18 in triplicates. We use iTaq DNA-polymerase in 20 ul PCR mix:
- 10 ul iTaq 2x Master Mix
- 2 ul Primer 10 uM each
- 6 ul water
- colony pick from plate
| Plasmid | Primer fw | Primer rv | Expected Fragment Size [bp] |
|---|---|---|---|
| pRB14 | VF2 | KH10 | 1000 |
| pRB15 | VF2 | RB36 | 1050 |
| pRB16 | VF2 | RB30 | 1100 |
| pRB17 | VF2 | RB34 | 1000 |
| pRB18 | VF2 | RB67 | 1050 |
Table 12.x
- PCR screening pRB14-18
| BioRAD T100 | ||
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 35 | 95 | 30 |
| 53 | 30 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
Annealing temperature according to NEB Tm calculator for Taq DNA polymerase and VF2
| Plasmid | Concentration [ng/ ul] | ~ size [bp] | Molarity [nM] |
|---|---|---|---|
| pRB15 | 98.7 | 3500 | |
| pRB16 | 106.6 | 3500 | |
| pRB17 | 188.3 | 3500 | |
| pRB18 | 129.9 | 3500 |
Test Transformation
OneShot and TOP10-sells have been transformed with pRB14. We expect OneShot cells to grow as usual and no colonies on TOP10. As a control group we transformed OneShot as well as TOP10 with pRB3.
pRB19
pRB19 is a pRB14-derived plasmid without the PPTase sfp. The genotype is pSB1C3-lacPromotor-BBa_B0034-indCdT(ccdB)
Table 12.x
- PCR Amplification of pRB14 for pRB19
| BioRAD T100 | ||
|---|---|---|
| RB21/46 from pRB14 miniprep 0.6 ng | ||
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 8 | 98 | 1 |
| TD 61 | 5 | |
| 72 | 100 | |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 100 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
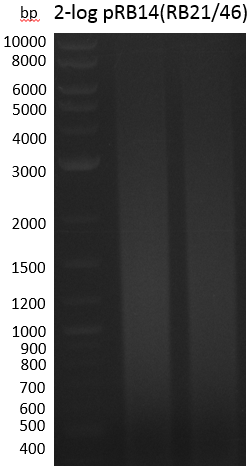
Smear -> Annealing temperatures in the touchdown-part were too high, so RB21 3' end couldn't bind properly. Second run with milder conditions.
Table 12.x
- PCR Amplification of pRB14 for pRB19 #2
| BioRAD T100 | ||
|---|---|---|
| RB21/46 from pRB14 miniprep 1 ng | ||
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| TD 60 | 5 | |
| 72 | 110 | |
| 25 | 98 | 1 |
| 65 | 5 | |
| 72 | 110 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
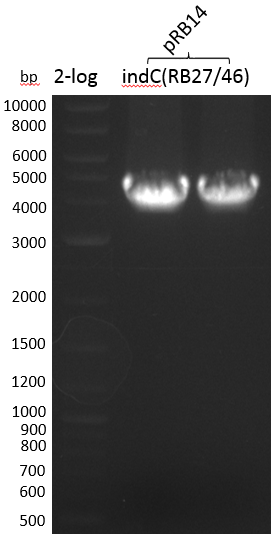
Smear -> we will run another strategy, i.e. amplification of indC from pRB14 with RB27/46 to further assemble it with pSB1C3(RB21/22)
Table 12.x
- PCR Amplification of indC(RB27/46) for pRB19 #3
| BioRAD T100 | ||
|---|---|---|
| RB27/46 from pRB14 miniprep 1 ng | ||
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| TD 57 | 5 | |
| 72 | 60 | |
| 25 | 98 | 1 |
| 65 | 5 | |
| 72 | 60 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
Gel extraction was performed using QIAquick gel extraction kit. DNA yield was measured using NanoDrop Spectrophotometer ND-1000
Table 12.x DNA concentrations for assembly of pRB19
| Fragment | Concentration [ng/ ul] | ~ Fragment size [bp] | Molarity [nM] |
|---|---|---|---|
| indC-pRB14(RB27/46) | 133.0 | 3800 | 53.03 |
| pSB1C3(RB21/22) | 210.8 | 2400 | 133.08 |
| Plasmid | Fragment 1 | Molarity [nM] | Volume in MM | |
|---|---|---|---|---|
| !Fragment 2 | Molarity [nM] | Volume in MM | ||
| pRB19 | indC-pRB14(RB27/46) | pRB14(RB21/46) | 53.03 | 3.3 |
| pSB1C3(RB21/22) | 133.08 | 1.7 |
Table 12.x
- CPEC Assembly pRB19
| BioRAD T100 | ||
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 53 | 5 | |
| 72 | 100 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
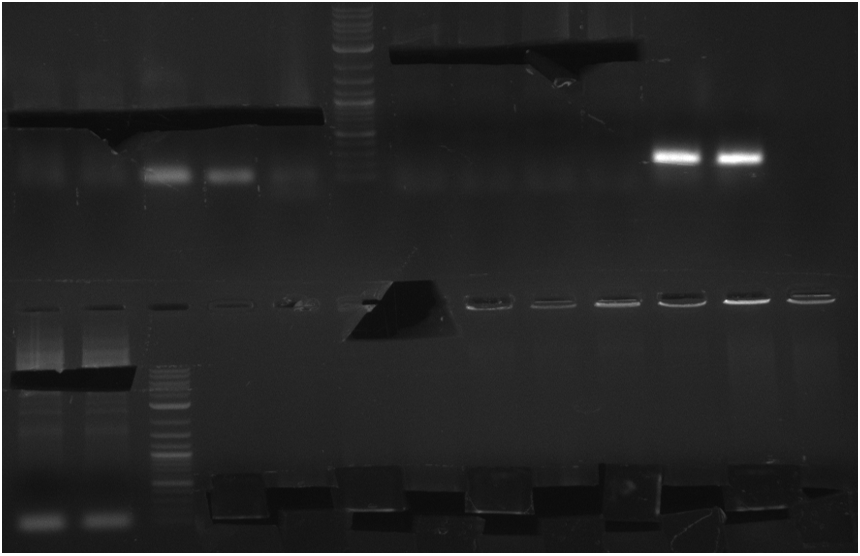
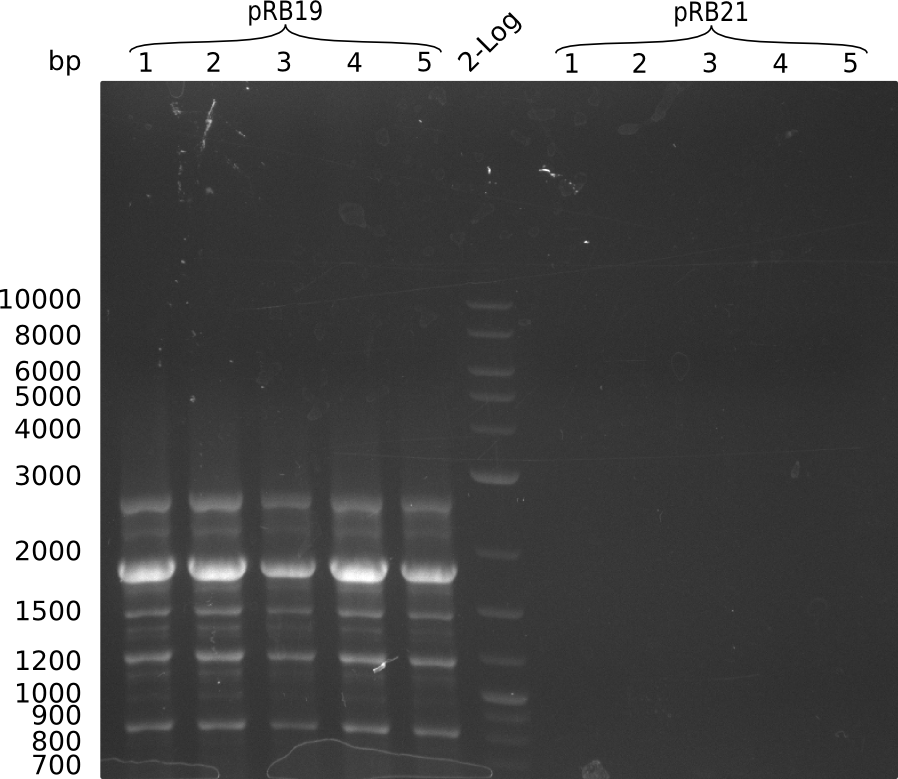
Transformation into OneShot Survival cells. Doing colony PCR for screening with primer KH9/VR which would give a fragment size of around 1.8 kbp: (Note: Throw picked colony 4 away since unintendently pooled 2 colonies in PCR tube). Used iTaq.
Table 12.x
- PCR screening pRB19 and pRB21
| Cycler 2 | ||
| KH9/VR from picked colonies. | ||
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 35 | 95 | 30 |
| 53 | 30 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
Colony PCR for pRB19 gave a lot of unspecific bands, probably because of less stringent parameters. Oculutated 6 ml of TB+Cm with colony 2 for MP preparation.
Biobricks
The PPTases sfp, svp, entD and delC and the indigoidine synthetase indC will be prepared for submission to the registry. indC contains two RFC10-cutting site, that have been removed during the assembly of pRB22.
pRB21 series
pRB21 is a pSB1C3-plasmid with lacPromotor, BBa_B0034 and indC. After the first assembly, EcoRI and SpeI cutting sites in indC will be removed to yield pRB22. After cutting site removal, the native T-Domain will be exchanged with ccdB to yield pRB22. pRB23 has the same genotype as pRB20 but with two PCR steps less.
Fragment Amplification
Table 12.x
- PCR Amplification of indC for pRB21 #1
| BioRAD T100 | ||
|---|---|---|
| RB27/46 from P. luminescens pellet | ||
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 8 | 98 | 1 |
| TD 61 | 5 | |
| 72 | 120 | |
| 25 | 98 | 1 |
| 65 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
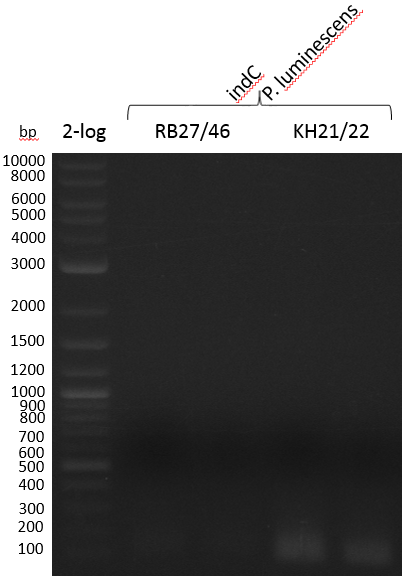
Amplification was not successful. Next try from PCR product with short primers as a rePCR.
Table 12.x
- PCR Amplification of indC for pRB21 #2
| BioRAD T100 | ||
|---|---|---|
| RB27/46 from indC short gel extraction | ||
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 8 | 98 | 1 |
| TD 56 | 5 | |
| 72 | 60 | |
| 25 | 98 | 1 |
| 65 | 5 | |
| 72 | 60 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
Heidelberg 20130815 PPTBBa indCpRB21 cut2.PNG
|
Gel Extraction
Gel extraction was performed using QIAquick gel extraction kit. DNA yield was measured using NanoDrop Spectrophotometer ND-1000
Table 12.x DNA concentrations for assembly of pRB21
| Fragment | Concentration [ng/ ul] | ~ Fragment size [bp] | Molarity [nM] |
|---|---|---|---|
| indC(RB27/46) | 63.7 | 3800 | 25.40 |
| pSB1C3(RB21/22) | 210.8 | 2400 | 133.08 |
CPEC Assembly and Transformation
| Plasmid | Fragment 1 | Molarity [nM] | Volume in MM | Fragment 2 | Molarity [nM] | Volume in MM |
|---|---|---|---|---|---|---|
| pRB21 | indC(RB27/46) | 25.40 | 3.5 | pSB1C3(RB21/22) | 133.08 | 1.5 |
Table 12.x
- CPEC Assembly pRB21
| BioRAD T100 | ||
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 53 | 5 | |
| 72 | 60 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
Validation
Colony PCR for pRB21 was unsuccessful (see gel picture), try again with less strigent parameters and positive control: KH5/VR on MP pRB3, which should give an amplicon size of around 2 kbp.
Table 12.x
- PCR screening pRB21
| T100 (right) | ||
| KH5/VR from picked colonies and 5 ng pRB3 MP | ||
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 35 | 95 | 30 |
| 50 | 30 | |
| 72 | 90 | |
| 1 | 72 | 150 |
| 1 | 12 | - |
Colony PCR for pRB21 worked now as well as the positive control. Oculutated 6 ml of TB+Cm with colony 8 for MP preparation.
PPTase BioBricks
have to look up used primer/template combinations again Konrad (talk)
Table 12.x
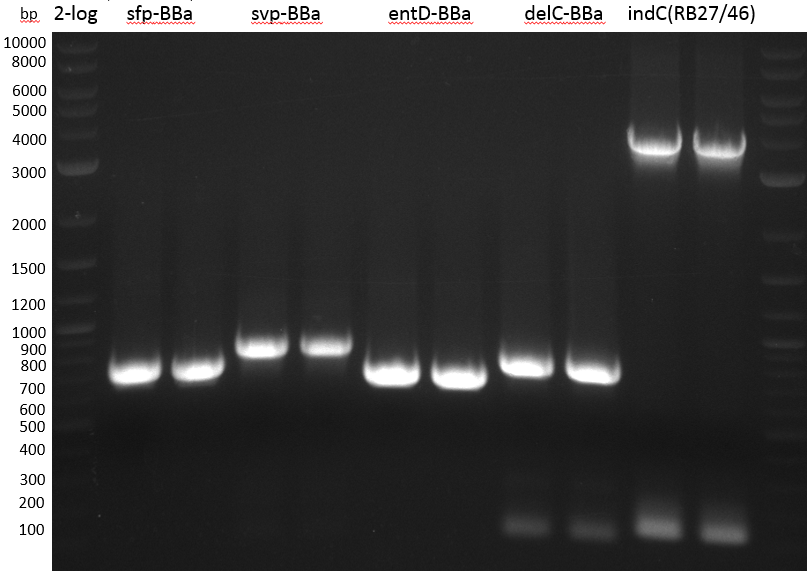
- PCR Amplification of PPTases for BioBrick submission
| BioRAD T100 | ||
|---|---|---|
| sfp: KH17/18 from pRB15 | ||
| svp: KH19/20 from pRB16 | ||
| entD: KH13/14 from pRB17 | ||
| delC: KH15/16 from pRB18 | ||
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| TD 66 | 5 | |
| 72 | 15 | |
| 25 | 98 | 1 |
| 65 | 5 | |
| 72 | 15 | |
| 1 | 72 | 60 |
| 1 | 12 | - |
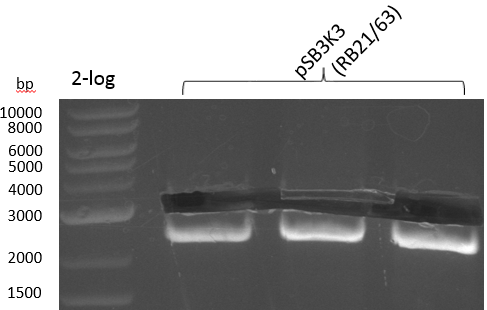
Heidelberg 20130815 PPTBBa indCpRB21 cut2.PNG
|
 "
"