Team:Heidelberg/Templates/MM week15p
From 2013.igem.org
Contents |
2013-08-05
- received constructs from Matthew Mattozzi, Boston: pET21c-pcc-acca-mcce in NEB Turbo (Ampr), pWH8: mcc-pcs-pcc-acc-mmce in NEB Turbo (Kanr), E. coli MM17: MG1655 lambda(DE3) d(lacY) d(E14) mcr-pcs-pcc-acc::KanR integrated at phage site HK022 (Kanr)
- plates contaminated with Drosophila larvae
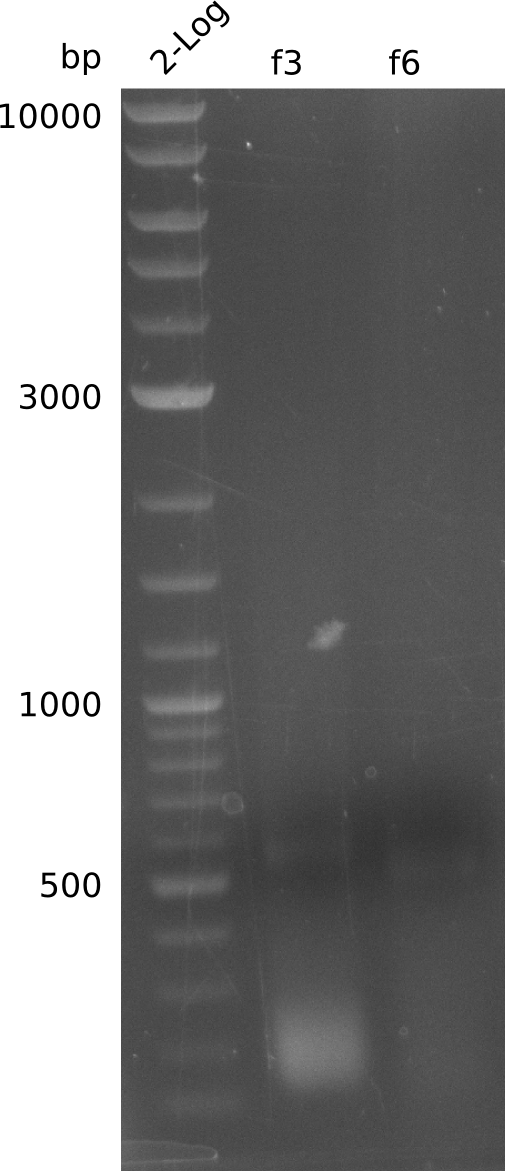
- re-streak on fresh plates, run colony-PCR for f3, f6 from pET21c-pcc-acca-mcce (Q5, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 35 | 98 | 5 |
| 61 | 10 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- no bands
2013-08-06
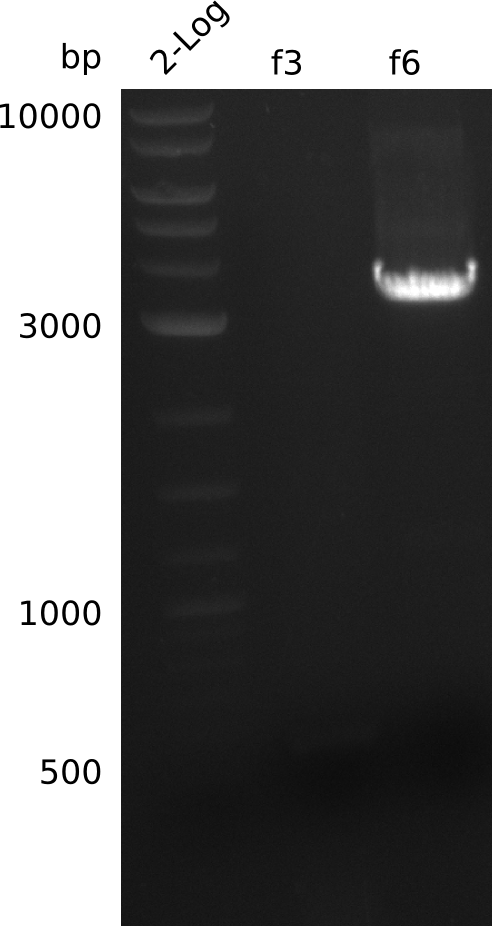
- run colony-PCR from pET21c-pcc-acca-mcce grown ON (Q5, 20 µl total volume) for fragments f3, f6:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 60 |
| 35 | 98 | 5 |
| 61 | 10 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- inoculate 5 ml 2xYT+Amp with colony from which colony-PCR was run, grow at 37°C
- no fragment for f3, re-run PCR (1 µl of liquid culture, 20 µl total, Phusion Flash):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 60 |
| 35 | 98 | 1 |
| 65 | 5 | |
| 72 | 60 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- make miniPrep of liquid culture
- no PCR product, miniPrep runs above 10 kb supercoiled (whole plasmid is expected to have 9 kb) (miniPrep has ca. 10 ng/µl)
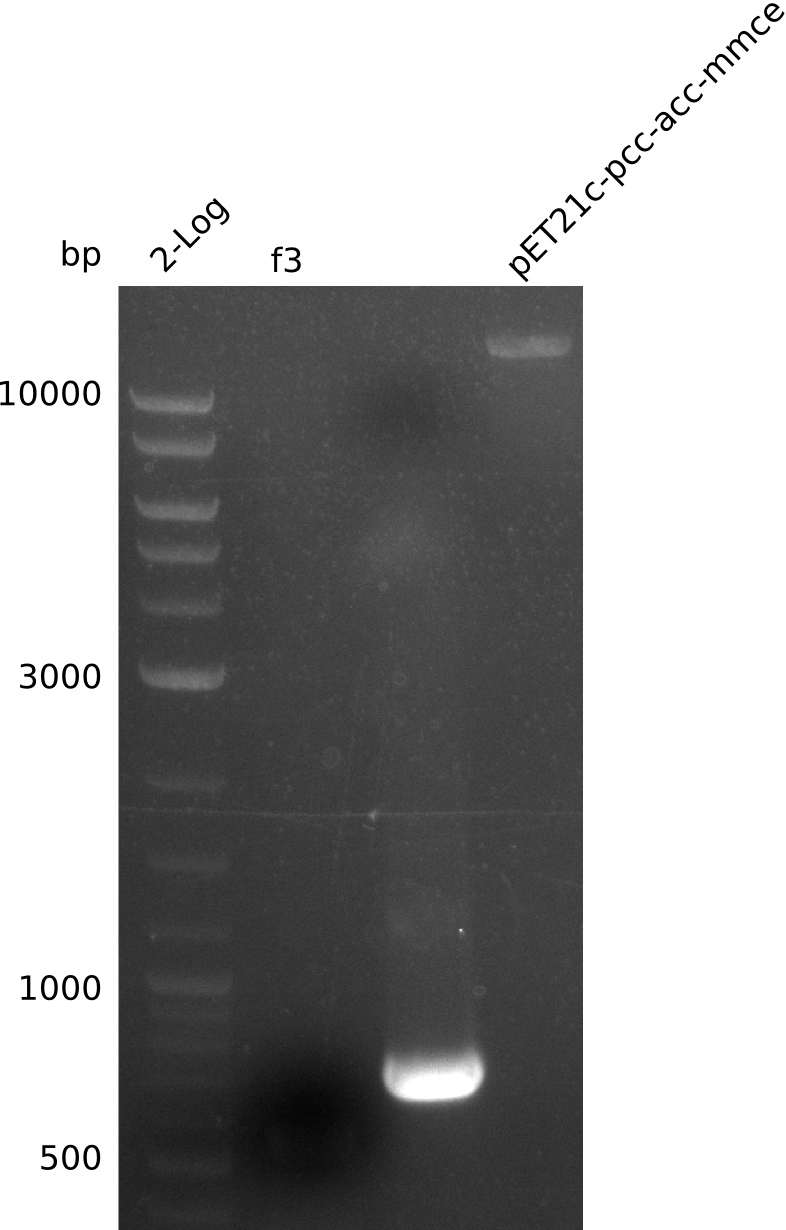
- re-run PCR for f3 from miniPrep (Q5, 0.2 µl DNA, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 60 |
| 35 | 98 | 5 |
| 61 | 10 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
2013-08-07
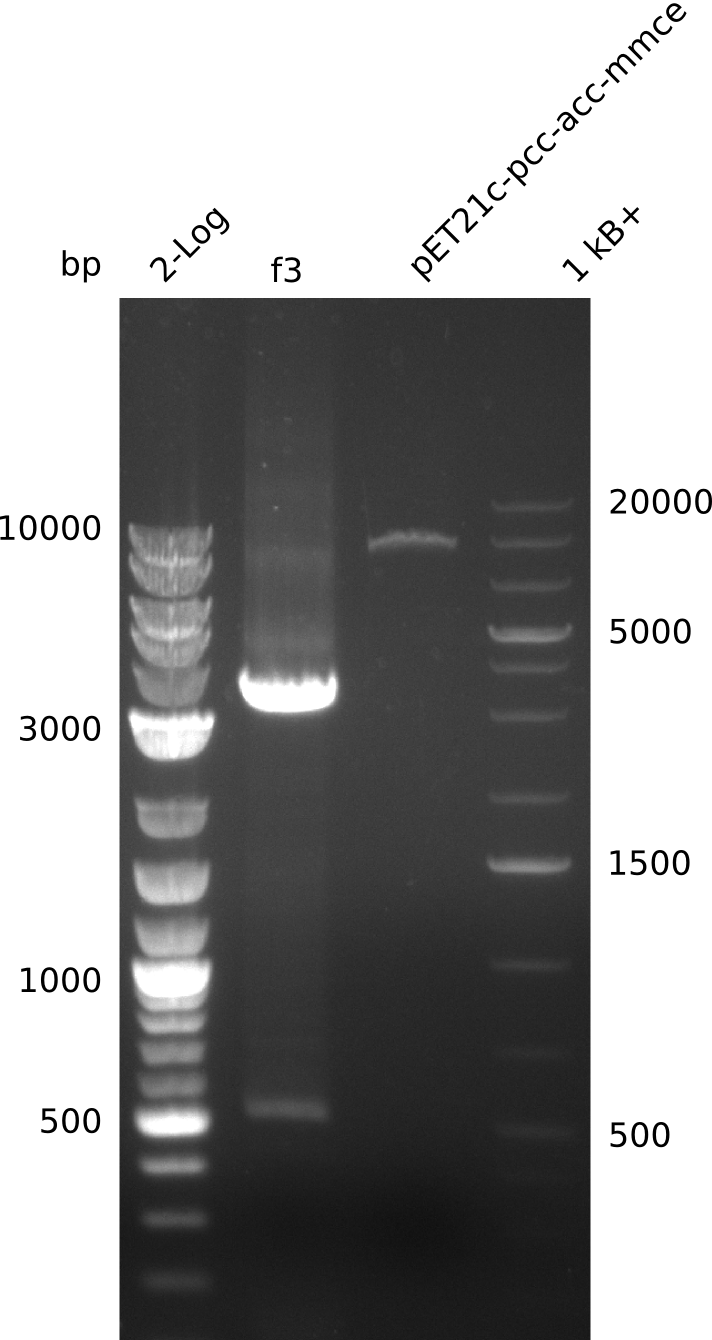
- need to verify plasmid identity: linearize with BamHI (unique restriction site in both pET21c... and pWH8, use 5 µl of miniPrep, 30 µl total volume)
- linearized pET21c... plasmid runs at 9 kb -> right
- PCR of f3 worked -> hypothesis: Gibson overhang against permeability device bound to E. coli genome in colony-PCRs, as permeability device is mutated version of FepA, which is present in E. coli
- gel-purify all fragments
- f1: 54.2 ng/µl, f2: 64.4 ng/µl, f3: 24.4 ng/µl, f4: 14 ng/µl, f5: 47.6 ng/µl, f6: 25.2 ng/µl; all in 30 µl
- perform Gibson assembly:
- pIK1: 0.8 µl f1, 1.6 µl f2, 5 µl f3, 2 µl f4, 0.4 µl H2O, 10 µl NEB Gibson MM
- pIK2: 1 µl f5, 6 µl f6, 3 µl f4
- incubate at 50°C for 1h
- transform TOP10: 2 transformations per plasmid: 5 µl of Gibson assembly, 5 µl of 1:4 diluted Gibson assembly
- grow in 2xYT at 37°C, plate on Cm, grow at 37°C
2013-08-08
- pIK1: several colonies, some of them pink -> pick 10 white, grow in 2xYT+Cm at 37°C
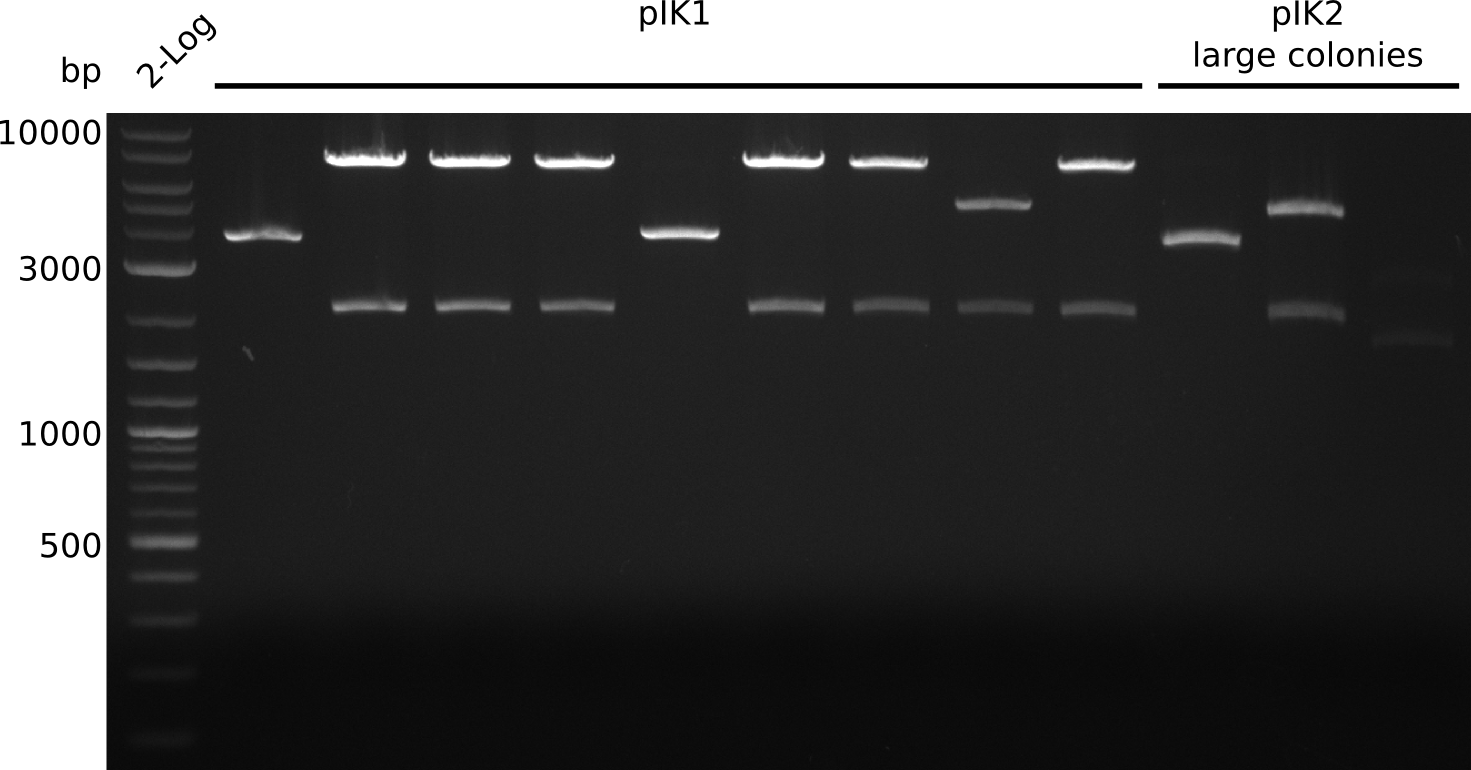
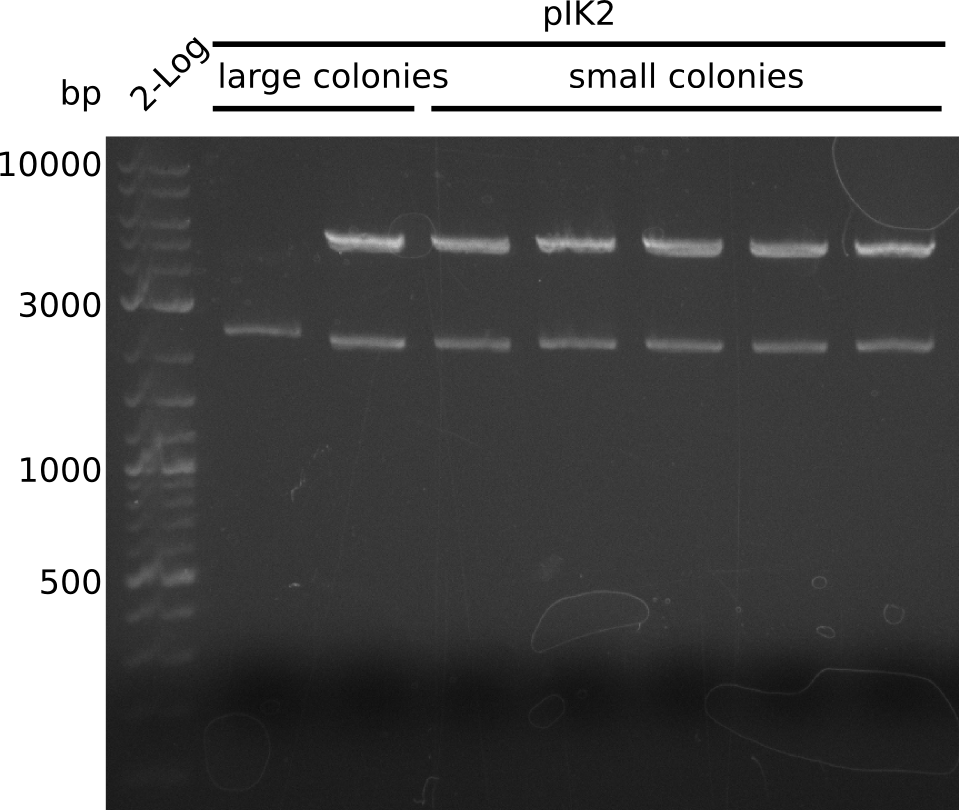
- pIK2: 2 populations of colonies: large and small, several large colonies are pink -> pick 5 white large, 5 white small colonies, grow in 3 ml 2xYT+Cm at 37°C
2013-08-09
- colony 8 of pIK1 did not grow
- make miniPreps of 2 ml everything else (2 ml each) -> ca. 62 - 600 ng/µl
- miniPrep yields quite high, but consistent: pACYC with p15A origin (4 kb) [http://www.abgent.com.cn/downloads/BG0003-Plasmid-mini-kit.pdf yields 0.6 µg DNA / ml culture] in LB, culture in 2xYT/TB yields 2-5 times more cells (according to QiaGen) => ca. 10-15 µg expected for 9 kb plasmid
- digest with BamHI+HindIII (20 µl total volume, 0.5 µl enzyme, 1 µl DNA [except for sample with 62 ng/µl, there 2 µl DNA was used])
- expected fragments: 2151 bp + 7265 bp for pIK1, 2151 bp + 5029 bp for pIK2
- send pIK1.2 and pIK2.2 to sequencing with primers VF2 and VR
 "
"