Team:Heidelberg/Tyrocidine week22 variation
From 2013.igem.org
Contents |
Gibson assembly of constructs 1 and 7
As these two constructs were the most divergent ones (construct 1 with only short linkers and construct 7 with only very long linkers) we decided to focus on these gibson assembly first This could help to to narrow functional regions.
| LV | Backbone-concentration | Ind-concentration | C4-concentration | C2-concentration |
|---|---|---|---|---|
| 1 | 3,27 µl | 4,42 µl | 1,76 µl | 0,55 µl |
| 7 | 0,97 µl | 5,26 µl | 2,42 µl | 1,34 µl |
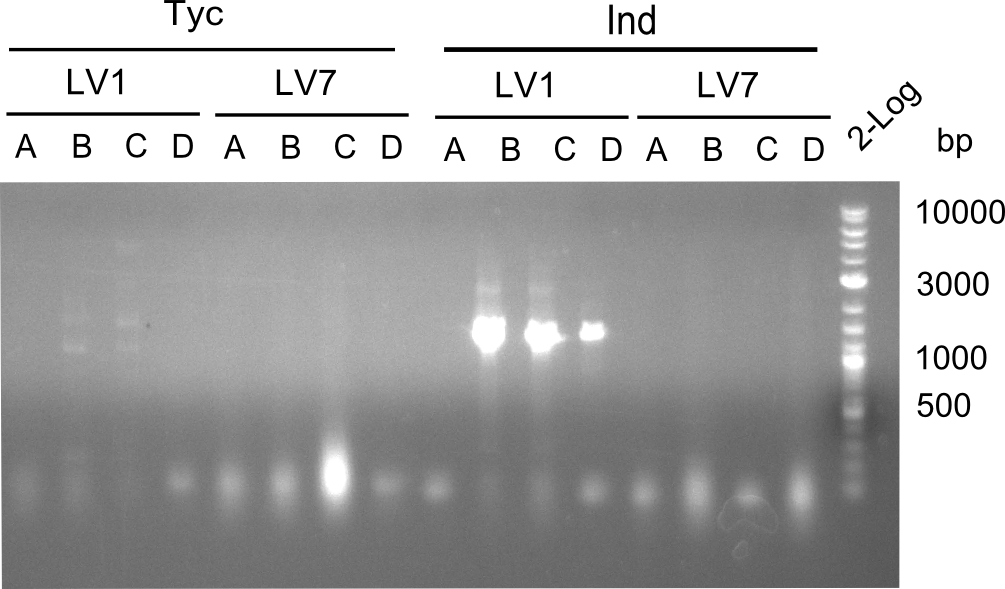
Afterwards,the constructs were transformed in DH10ß-cells and platet on LB-plates with chloramphenicol. The day after, the Plates containing LV1 and LV7 showed colonies. Therefore colony PCRs were performed.
Mastermixes for colony PCRs:
| What | µl |
|---|---|
| iTaq-Polymerase | 50 |
| H2O | 48 |
| Primer VF2 | 1 |
| Primer PW20 | 1 |
Mastermix for the indC Amplification:
| What | µl |
|---|---|
| iTaq-Polymerase | 50 |
| H2O | 48 |
| Primer VR | 1 |
| Primer KH5 | 1 |
PCR conditions:
| Biometra T100 | ||
| Cycles | temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 35 | 95 | 0:30 |
| 52 | 0:30 | |
| 72 | 2:30 | |
| 1 | 72 | 5:00 |
| 1 | 10 | inf |
Results
Only for construct LV1C a slight band for the Tyrocidine module was visble. To make sure, that there was no mistake in the colony PCR itself, we decided to transform every LV1 construct in BAP. Overnight cultures of them were minipreped with a QiaPrep-Kit.
Concentrations measured via NanoDrop:
| Construct | Concentration in ng/µl | A260/280 |
|---|---|---|
| LV1A | 10,6 | 0,211/0,1 |
| LV1B | 12,2 | 0,244/0,104 |
| LV1C | 11,3 | 0,226/0,113 |
| LV1D | 3,7 | 0,073/-0,017 |
Chemical Transformation of LV1 constructs in BAPI
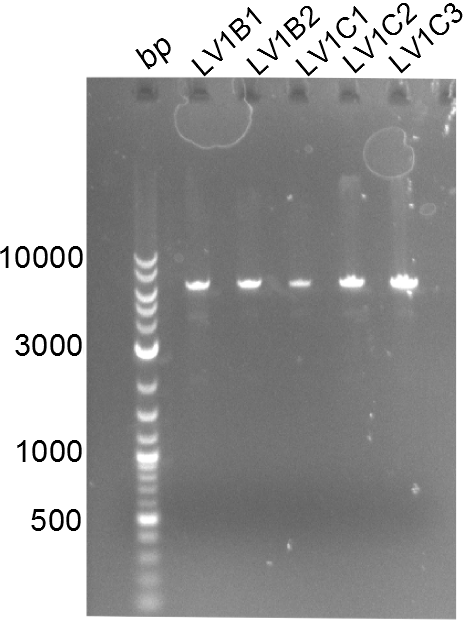
Positive constructs were transformed in BAPI-cells and spread on plates. There were slight blue circles around the colonies of LV1B and LV1C. Colonies of all plates were picked. Without induction LV1B1, LV1B2, LV1B3, LV1C1, LV1C3 turned into a clear visible dark blue. 2ml of these culture were minipreped and treated by restriction digested with ecoR1.
| What | µl |
|---|---|
| CutSmart | 2 |
| EcoR1 | 1 |
| DNA | 3 |
| ddH2O | 14 |
Results
All LV1 constructs were negative. The Gibson Transformation has to be done again.
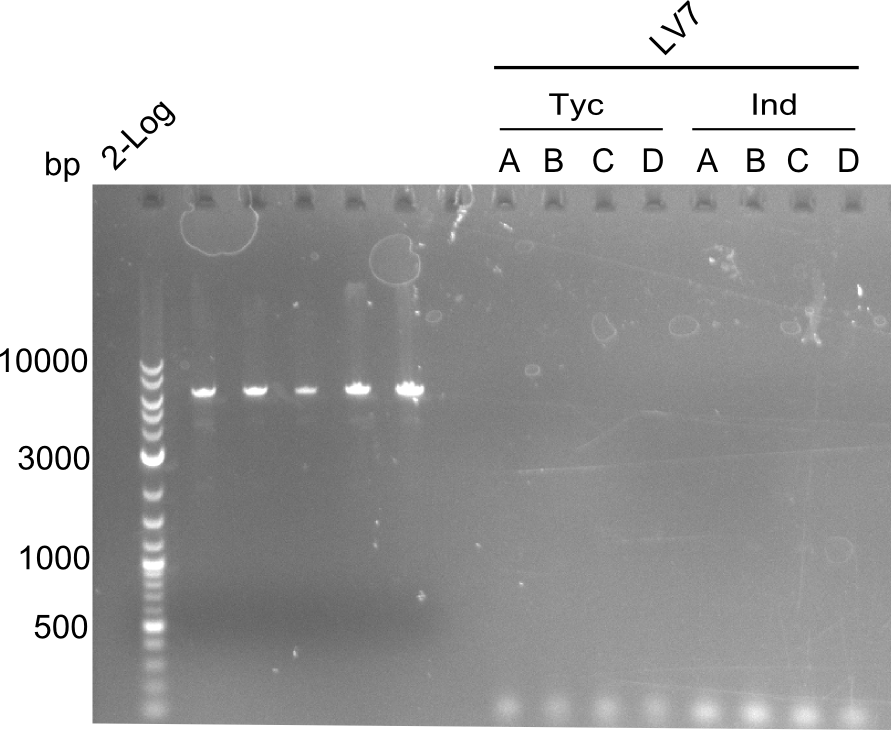
colony PCR of LV7
As the former colony-PCR was negative, again colonies of DH10ß were picked and a colony PCR redone.
Mastermix for TycC4dC-C(TycC2) amplification:
| What | µl |
|---|---|
| iTaq-Polymerase | 5 |
| H2O | 3 |
| Primer VF2 | 1 |
| Primer PW20 | 1 |
Mastermix for the Indigoidine Amplification
| What | µl |
|---|---|
| iTaq-Polymerase | 5 |
| H2O | 3 |
| Primer VR | 1 |
| Primer KH5 | 1 |
Conditions
| Biometra T100 | ||
| Cycles | Temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 35 | 95 | 0:30 |
| 52 | 0:30 | |
| 72 | 2:30 | |
| 1 | 72 | 5:00 |
| 1 | 10 | inf |
Gibson assembly of the other linker-variation constructs
As we had putative positive clones for LV1, we did a Gibson-assembly with the other constucts, too.
Mastermix for Gibson Assembly
10µl Gibson-mastermix + LV-Mix
| LV | BB | indC | TycC4dC | C(TycC2) |
|---|---|---|---|---|
| 2 | 0,75 (short) | 5,22 (medium) | 1,62 (short) | 2,41 (medium) |
| 3 | 1,59 (medium) | 3,23 (short) | 4,78 (medium) | 0,41 (short) |
| 5 | 1,54 (medium) | 3,12 (long) | 4,62 (medium) | 0,71 (long) |
| 6 | 0,81 (long) | 4,36 (medium) | 2,25 (long) | 2,58 (medium) |
| 8 | 3,09 (short) | 4,18 (long) | 1,67 (short) | 1,08 (long) |
| 9 | 1,01 (long) | 5,48 (short) | 2,82 (short) | 0,68 (short) |
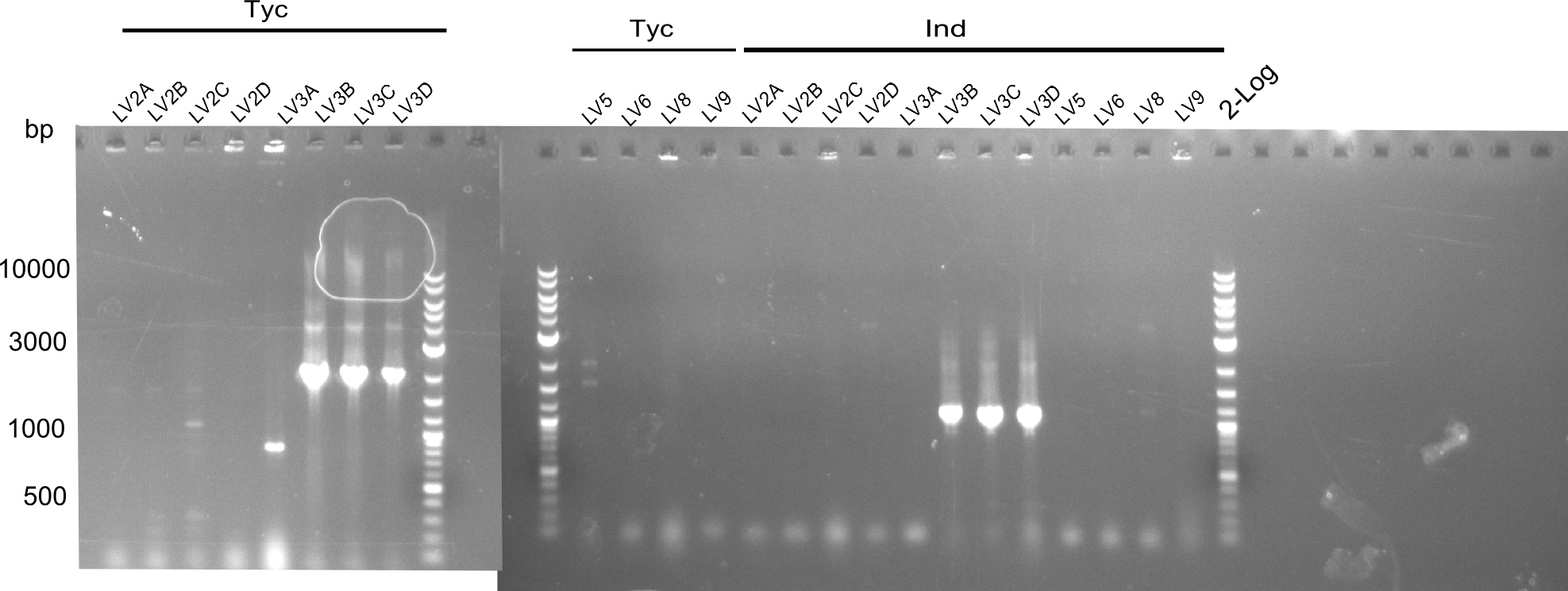
These mixes were incubated at 50°C for 1h. Afterwards they were transformed by electroporation and plated out. Colonies were picked and a colony PCR was executed.
Mastermix for TycC4dC-C(TycC2) amplification
| What | µl |
|---|---|
| iTaq-Polymerase | 5 |
| H2O | 3 |
| Primer VF2 | 1 |
| Primer PW20 | 1 |
Mastermix for the Indigoidine amplification
| What | µl |
|---|---|
| iTaq-Polymerase | 5 |
| H2O | 3 |
| Primer VR | 1 |
| Primer KH5 | 1 |
Conditions
| Biometra T100 | ||
| Cycles | temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 35 | 95 | 0:30 |
| 52 | 0:30 | |
| 72 | 2:30 | |
| 1 | 72 | 5:00 |
| 1 | 10 | inf |
The constructs of LV3b,c,d were positive, so the liquid cultures were prepared.
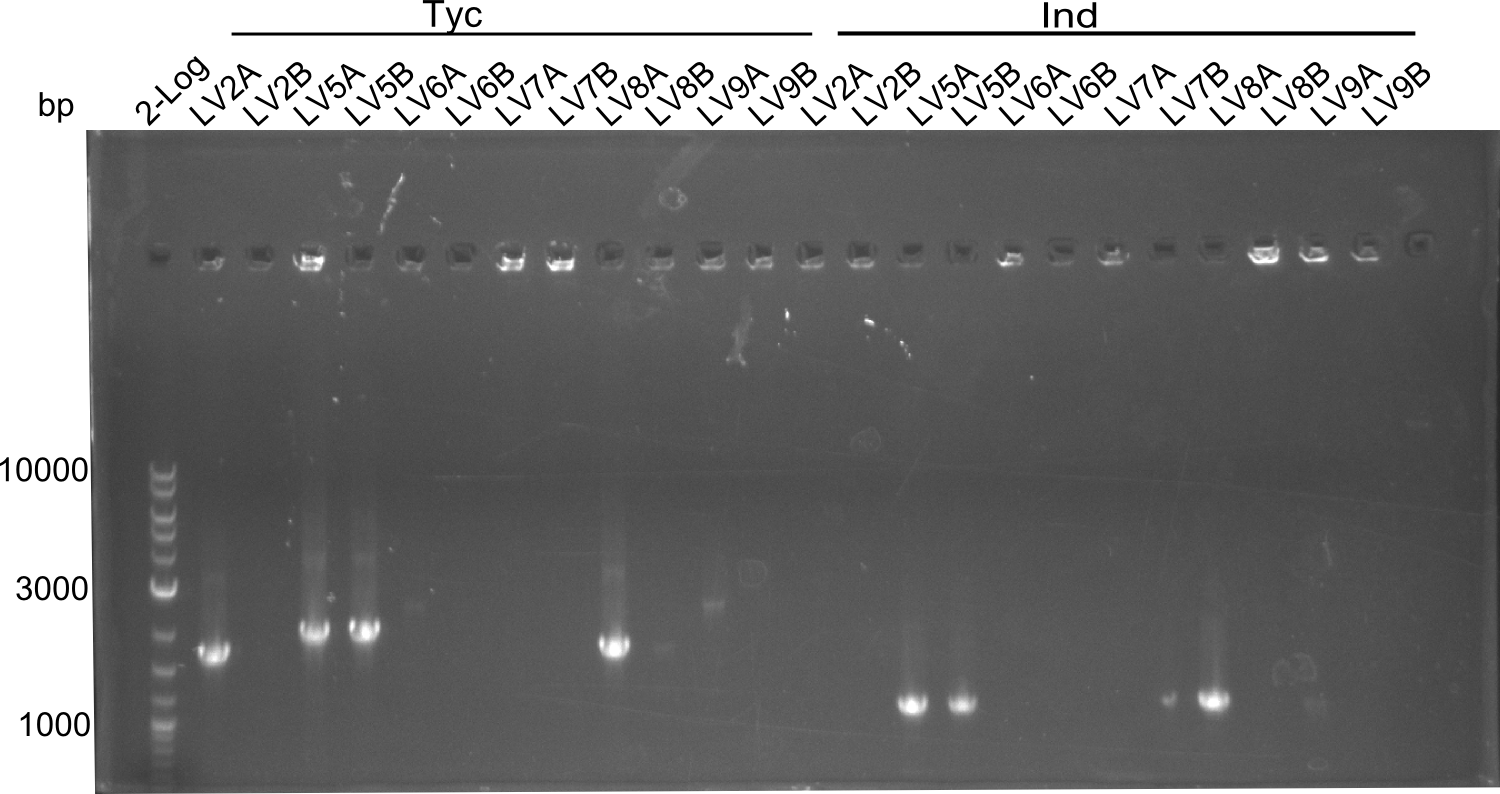
colony PCR of LV 2,5,6,7,8,9
| What | µl |
|---|---|
| oneTaq-Polymerase | 5 |
| H2O | 3 |
| Primer VF2 | 1 |
| Primer PW20 | 1 |
Mastermix for the Indigoidine Amplification
| What | µl |
|---|---|
| oneTaq-Polymerase | 5 |
| H2O | 3 |
| Primer VR | 1 |
| Primer KH5 | 1 |
Conditions
| Biometra T100 | ||
| Cycles | temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 35 | 95 | 0:30 |
| 52 | 0:30 | |
| 72 | 2:30 | |
| 1 | 72 | 5:00 |
| 1 | 10 | inf |
 "
"