Team:Nevada/project/OMpermeabilization
From 2013.igem.org


Improvement of currently registered part BBa_K523013
Background
The plant pathogen, Pseudomonas syringae, expresses a unique outer membrane surface protein which is used by the pathogen to promote ice formation (Orser et al., 1985) (Wolber et al., 1986) (Deininger et al., 1988). This protein, called an ice nucleation protein (INP), has been taken advantage of by various researchers as a cell surface display model when fused with a florescent protein such as yellow fluorescent protein (YFP) (Lindow et al., 1989). An alarming aspect of this protein in regards to practical research applications is its size at. In Li et. Al (2004) (Li et al., 2004) (Gurian-Sherman and Lindow, 1993) (, a truncated INP with its C-terminal and internal repeats removed (INP-N) was fused with GFP and showed improved rates of outer membrane localization compared to full length proteins (~1400 amino acids) or ones with simply the internal repeats removed (INP-NC) (Li et al., 2004). In 2011, Edinburgh took the approach of using an INP-NC protein within the iGEM registry (BBa_K265008) and fusing it to YFP in constructing a cell surface display.
Our Approach
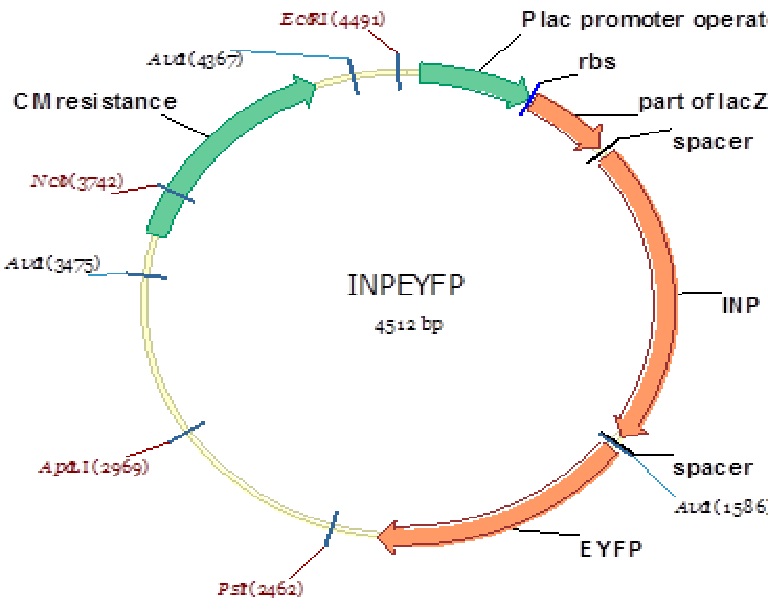
Our project this year was concerned with developing a novel bactericide for gram negative bacteria. Since our bactericide would be used to lyse the peptidoglycan layer of these pathogens from the external environment (Kadurugamuwa and Beveridge, 1996), we needed to develop an assay to examine the efficacy of outer membrane (OM) permeabilization techniques which would be coupled with our bactericide. We chose to develop an assay using an outer membrane florescent cell surface detection method with the theory that any OM which was permabilized would pellet following centrifugation post exposure to the given chemical of interest. We would then quantify the results by examining the intensity of the resultant pellet. Therefore, we needed to develop a cell surface detection protein to be expressed in our gram negative bacteria model, E. coli. We knew many of these proteins existed in the iGEM registry so we aimed to find one, then improve upon it. One of Edinburgh’s parts from 2011 looked promising for our needs. This part was a fusion of an ice nucleation protein (INP) and a yellow fluorescent protein (YFP). Per Edinburghs 2011 iGEM wiki page, their part consisted of a LacZ promoter (Plac), the N and C terminal domains of INP, and YFP (Plac-INPNC-YFP). Per Li et. al (2004), we decided to improve this part by deleting the C-terminal domain of the INP through site directed mutagenesis. The final construct, Plac-INPN-YFP was constructed in E. coli.
Design 1
Given the efficacy of the protein, as described by Edinburgh, we chose to do site directed mutagenesis on the gene while in pSB1C3 and keep it under the control of the LacZ promoter. Further investigation revealed that the original part used by Edinburgh did not have the entire internal sequence deleted; we therefore took the approach of designing primers that would yield a more efficient protein containing just the LacZ promoter region, the INP N-terminal, and YFP. Edinburgh’s design was 2442 base pairs in size. We deleted 372 base pairs yielding a 2070 base pair construct.
- Primers were designed based on Gibson Assembly protocols and the internal sequence to be deleted so as to keep the protein in pSB1C3 while we were working with it.
- Gibson assembly was carried out
- PCR followed by gel electrophoresis revealed that we had in fact deleted the appropriate length
- Sequencing data revealed that we had deleted the appropriate region
- This construct was transformed into BL21 cells and induced with 100mM IPTG
Results: This design resulted in the deletion of 372 base pairs yielding a 2070 base pair construct of Plac-INPN-YFP as evidenced by gel electrophoresis (figure 10). Fluorescence observed under fluorescent microscopy at equivalence to Edinburgh’s construct.
Figure 9: Plca-INPN-YFP Plasmid map
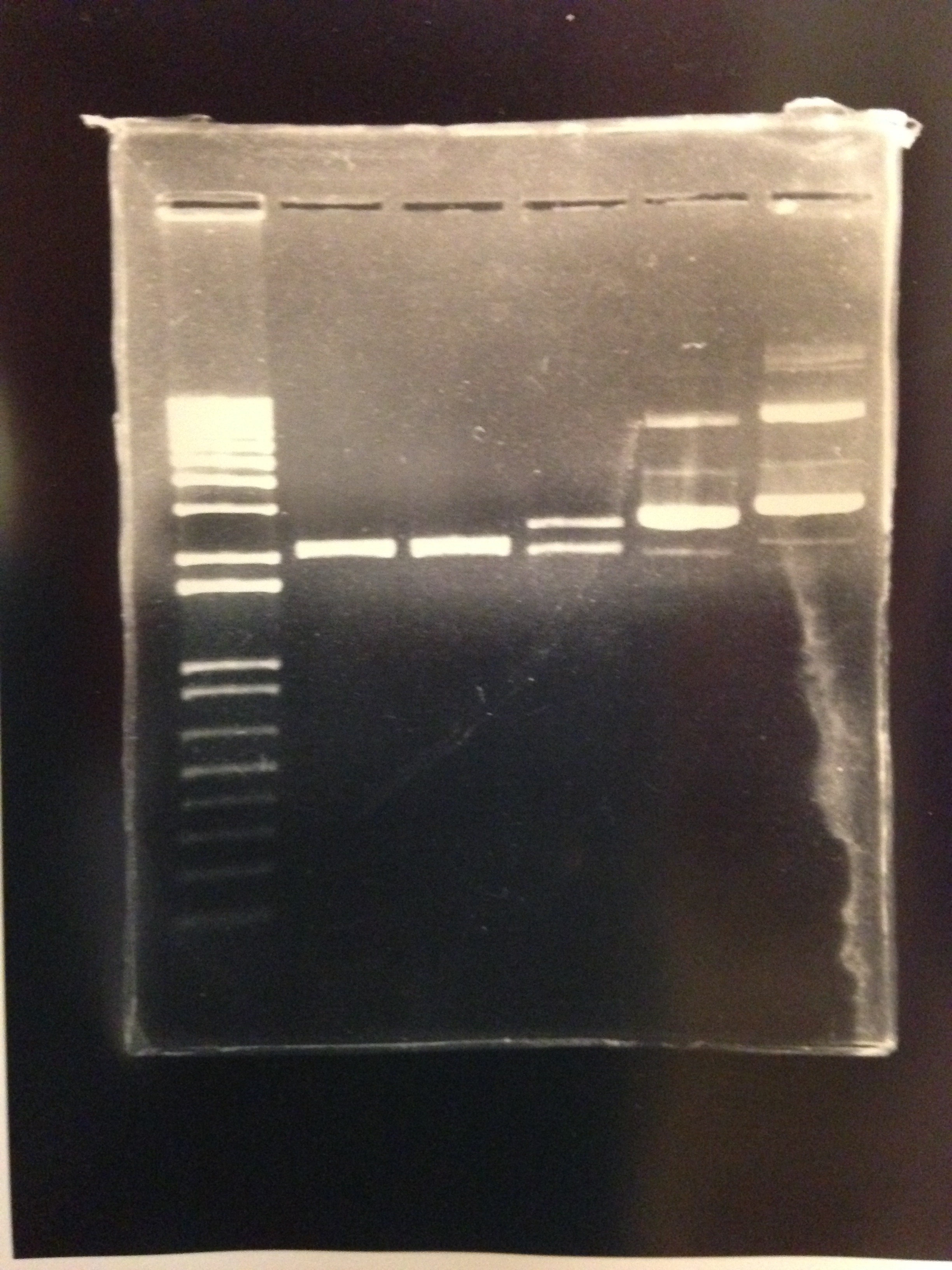
Figure 10: 1% agarose of PstI and EcorI digested Plac-INPN-YFP and Plac-INPNC-YFP: Lane 1 is a 1kb marker. Lanes 2 and 3 are PstI and EcoRI digested pSB1C3 with expected construct at 2070 bp. pSB1C3 is also 2070 bp therefore the correct construct will show as one band with the backbone. Lane 4 is PstI and EcoRI digested pSB1C3 with the original Plac-INPNC-YFP construct. Lanes 5 and 6 are undigested Plac-INPN-YFP and Plac-INPNC-YFP respectfully.
Design 2
Given lack of significance displayed by the first model, we spent some time looking at the original design and noticed a large region between the LacZ promoter and the INP N-terminal which was composed of a truncated, and non-functional, LacZ gene. This coupled with our speculation that there may be a mutation in the promoter itself, lead us to move the sequence into the plasmid pBAD under the control of an arabinose induced promoter. Design 1 was used as the template for this design and yielded construct INPN-YFP.
- Primers were designed based on the desired deletion of the LacZ promoter and truncated LacZ gene.
- Our pBAD vector was transformed into LMG 194 cells
- Induction with arabinose
Results
This design resulted in the deletion of 622 base pairs yielding a 1448 base pair construct of INP-N-YFP as evidenced by gel electrophoresis (figure 11). No fluorescence was detected in this design.
Figure 11: 1% agarose of PCR amplified INPN-YFP in lane 2 against a 1 kb marker in lane 1. Expected band size of 1448 observed.
 "
"