From 2013.igem.org
Aug 20
Miniprep
Kojima
| DNA | concentration[µg/µL] | 260/280 | 260/230
|
| 8/18 Ptet -1 | 220 | 1.69 | 1.87
|
| 8/18 Ptet -2 | 210 | 1.69 | 1.71
|
| 8/18 RBS-tetR-DT -1 | 236 | 1.63 | 1.69
|
| 8/18 RBS-tetR-DT -2 | 182 | 1.65 | 1.98
|
| 8/18 J23100-RBS-GFP-DT -1 | 334 | 1.71 | 2.12
|
| 8/18 J23100-RBS-luxR-DT -1 | 440 | 1.67 | 1.60
|
| 8/18 J23100-RBS-luxR-DT -2 | 344 | 1.68 | 1.83
|
| 8/18 J23100-RBS-lacZα-DT -1 | 280 | 1.70 | 1.81
|
| 8/18 RBS-lysis3 -1 | 282 | 1.67 | 1.76
|
| 8/18 RBS-lysis3 -2 | 212 | 1.30 | 1.35
|
LB Medium Plate
Okazaki
| volume | 200mL
|
| Bacto Trypton | 2g
|
| Bacto yeast extract | 1g
|
| Nacl | 1g
|
| Agar Powder | 2g
|
| Amp | 40µl
|
1. Measuring reagents and put they in the Erlenmeyer flask.
2. diluting in measuring cylinder to 200ml total.
3. autoclaving.
4. adding ampicirine 40ml.
Ristriction Enzyme Digestion
Kojima and Ashida
| | 8/20 Ptet -1(220µg/mL) | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 4.5 | 1 | 1 | 3 | 3 | 17.5 | 30
|
| 1 cut | 0.5 | 0.2 | 0 | 1 | 1 | 7.3 | 10
|
| 1 cut | 0.5 | 0 | 0.2 | 1 | 1 | 7.3 | 10
|
| NC | 0.5 | 0 | 0 | 1 | 1 | 7.5 | 10
|
| | 8/20 RBS-tetR-DT -2(182µg/mL) | EcoRI | SpeI | 10x buffer H | MilliQ | total
|
| 2 cuts | 5.5 | 1 | 1 | 3 | 19.5 | 30
|
| 1 cut | 0.6 | 0.2 | 0 | 1 | 8.2 | 10
|
| 1 cut | 0.6 | 0 | 0.2 | 1 | 8.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 8.4 | 10
|
| | 8/20 J23100-RBS-GFP-DT(334µg/ml) | EcoRI | SpeI | 10x buffer H | MilliQ | total
|
| 2 cuts | 3.0 | 1 | 1 | 3 | 22 | 30
|
| 1 cut | 0.3 | 0.2 | 0 | 1 | 8.5 | 10
|
| 1 cut | 0.3 | 0 | 0.2 | 1 | 8.5 | 10
|
| NC | 0.3 | 0 | 0 | 1 | 8.7 | 10
|
| | 8/20 J23100-RBS-luxR-DT -2(344µg/mL) | EcoRI | XbaI | BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 2.9 | 1 | 1 | 3 | 3 | 19.1 | 30
|
| 1 cut | 0.3 | 0.2 | 0 | 1 | 1 | 7.5 | 10
|
| 1 cut | 0.3 | 0 | 0.2 | 1 | 1 | 7.5 | 10
|
| NC | 0.3 | 0 | 0 | 1 | 1 | 7.7 | 10
|
| | 8/20 RBS-lysis3 -1(282µg/mL) | EcoRI | SpeI | 10x buffer H | MilliQ | total
|
| 2 cuts | 3.5 | 1 | 1 | 3 | 21.5 | 30
|
| 1 cut | 0.4 | 0.2 | 0 | 1 | 8.4 | 10
|
| 1 cut | 0.4 | 0 | 0.2 | 1 | 8.4 | 10
|
| NC | 0.4 | 0 | 0 | 1 | 8.6 | 10
|
| | 8/17 DT -1(188µg/mL) | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 5.3 | 1 | 1 | 3 | 3 | 16.7 | 30
|
| 1 cut | 0.3 | 0.2 | 0 | 1 | 1 | 7.5 | 10
|
| 1 cut | 0.3 | 0 | 0.2 | 1 | 1 | 7.5 | 10
|
| NC | 0.3 | 0 | 0 | 1 | 1 | 7.7 | 10
|
Transformation
Nakamoto
| Name | Sample | Competent Cells(XL10-gold) | Total | Plate
|
| tRNA-Spinach-tRNA | 1µL | 10µL | 11µL | Amp
|
| tetR-aptamer 12_PC(076) | 1µL | 10µL | 11µL | Amp
|
| tetR-aptamer 12_1R(113) | 1µL | 10µL | 11µL | Amp
|
| tetR-aptamer 12_1M(105) | 1µL | 10µL | 11µL | Amp
|
| pT181 attenuator | 1µL | 10µL | 11µL | Amp
|
| Fusin1 attenuator | 1µL | 10µL | 11µL | Amp
|
| Fusion3m2 attenuator | 1µL | 10µL | 11µL | Amp
|
| pT181 antisense | 1µL | 10µL | 11µL | Amp
|
| Fusion1 antisense | 1µL | 10µL | 11µL | Amp
|
| Fusion6 antisense | 1µL | 10µL | 11µL | Amp
|
incubate 37°C overnight 20:34~
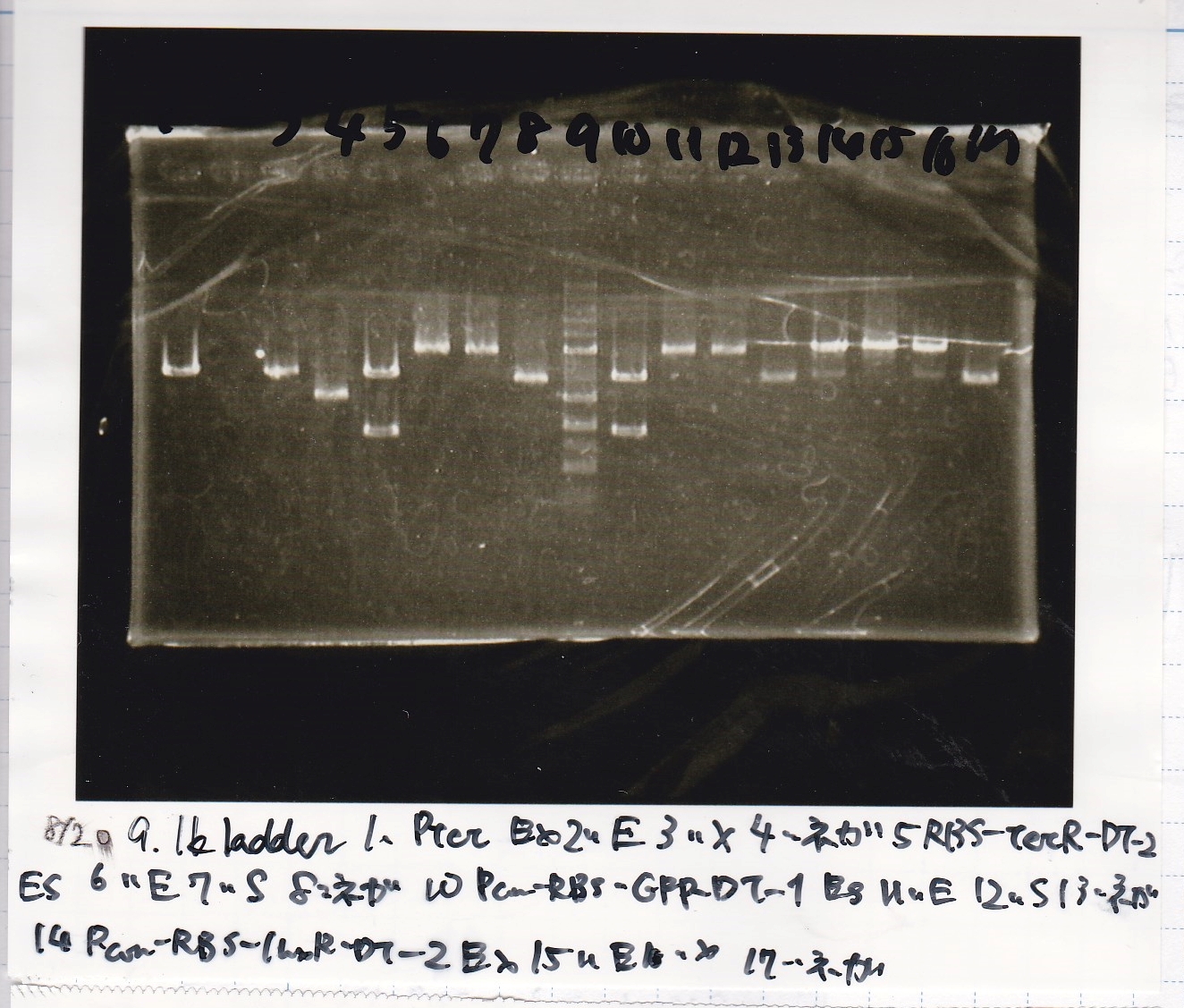
Electrophoresis
Kojima
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 8/20 Ptet -1 | EcoRI | XbaI
|
| 2 | 8/20 Ptet -1 | EcoRI | --
|
| 3 | 8/20 Ptet -1 | -- | XbaI
|
| 4 | 8/20 Ptet -1 | -- | --
|
| 5 | 8/20 RBS-tetR-DT -2 | EcoRI | SpeI
|
| 6 | 8/20 RBS-tetR-DT -2 | EcoRI | --
|
| 7 | 8/20 RBS-tetR-DT -2 | -- | SpeI
|
| 8 | 8/20 RBS-tetR-DT -2 | -- | --
|
| 9 | 1kbp ladder | -- | --
|
| 10 | 8/20 J23100-RBS-GFP-DT -1 | EcoRI | SpeI
|
| 11 | 8/20 J23100-RBS-GFP-DT -1 | EcoRI | --
|
| 12 | 8/20 J23100-RBS-GFP-DT -1 | -- | SpeI
|
| 13 | 8/20 J23100-RBS-GFP-DT -1 | -- | --
|
| 14 | 8/20 J23100-RBS-luxR-DT -1 | EcoRI | XbaI
|
| 15 | 8/20 J23100-RBS-luxR-DT -1 | EcoRI | --
|
| 16 | 8/20 J23100-RBS-luxR-DT -1 | -- | XbaI
|
| 17 | 8/20 J23100-RBS-luxR-DT -1 | -- | --
|
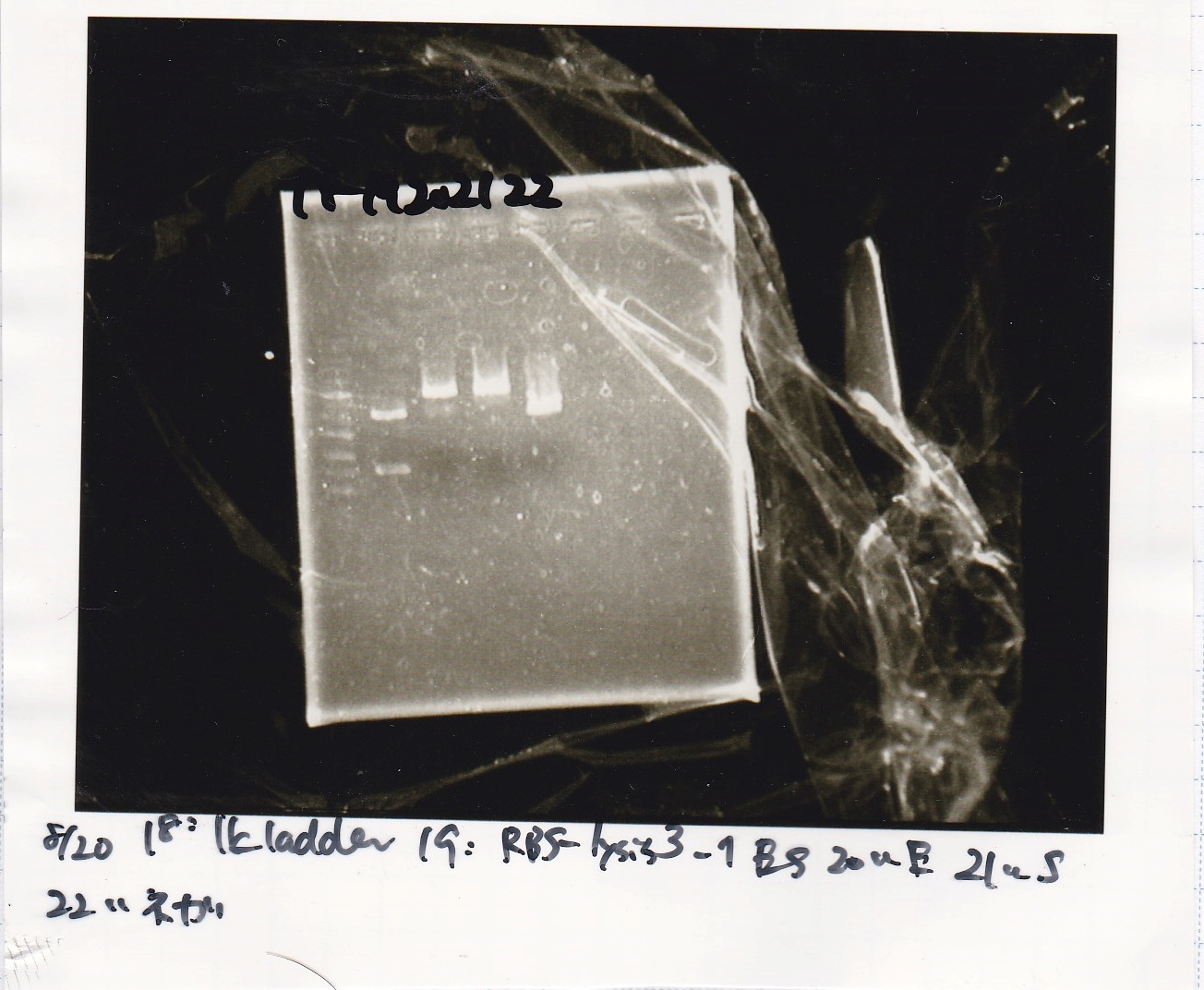
| 18 | 1kbp ladder | -- | --
|
| 19 | 8/20 RBS-lysis3 -1 | EcoRI | SpeI
|
| 20 | 8/20 RBS-lysis3 -1 | EcoRI | --
|
| 21 | 8/20 RBS-lysis3 -1 | -- | SpeI
|
| 22 | 8/20 RBS-lysis3 -1 | -- | --
|
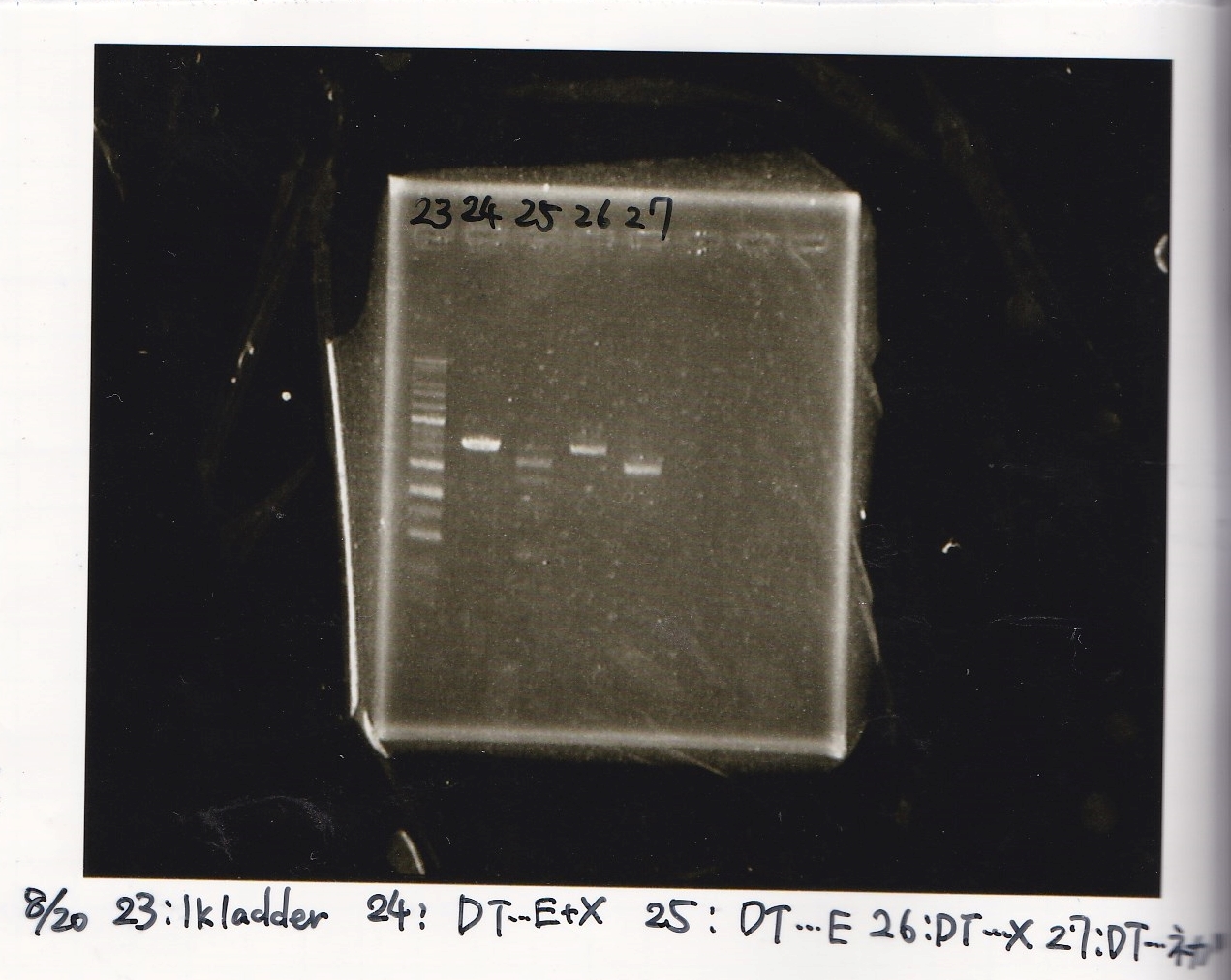
| 23 | 1kbp ladder | -- | --
|
| 24 | 8/17 DT | EcoRI | XbaI
|
| 25 | 8/17 DT | EcoRI | --
|
| 26 | 8/17 DT | -- | XbaI
|
| 27 | 8/17 DT | -- | --
|
 "
"


