Team:BYU Provo/Notebook/SmallPhage/Summer/Period1/Exp/7.13 Phage Propagation
From 2013.igem.org
(Difference between revisions)
| (6 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{| width="100%" | {| width="100%" | ||
| - | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> '''Small Phage | + | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> '''Small Phage July - August Notebook: Experiments'''</font> |
<br> | <br> | ||
| Line 14: | Line 14: | ||
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | : | + | <font size = "4"> |
| + | |||
| + | : <u> '''Small Phage''' </u> </font> | ||
: [[Team:BYU Provo/Notebook/SmallPhage/Winterexp|March-April]] | : [[Team:BYU Provo/Notebook/SmallPhage/Winterexp|March-April]] | ||
| Line 45: | Line 47: | ||
'''III) Reagent Record''' | '''III) Reagent Record''' | ||
| - | : Chloroform from Dr. Grose’s lab; LB; E coli BL21 liquid culture overnight; x8 top agar; 5.3 phage stock | + | : Chloroform from Dr. Grose’s lab; LB; E coli BL21 liquid culture overnight; x8 top agar; x2 top agar; 5.3 phage stock |
<br> | <br> | ||
| Line 57: | Line 59: | ||
:: :1mL phage: 4mL LB + 1mL E coli + 1mL 5.3 phage stock | :: :1mL phage: 4mL LB + 1mL E coli + 1mL 5.3 phage stock | ||
| - | : 2. Liquid culture purification | + | : 2. Liquid culture purification 1(7.15) |
:: 1) For each infected test tubes (10uL, 100uL, 1mL), put 1mL of liquid culture into 5 eppendorf tubes. | :: 1) For each infected test tubes (10uL, 100uL, 1mL), put 1mL of liquid culture into 5 eppendorf tubes. | ||
:: 2) Centrifuge at 3000 rpm for 5 minutes | :: 2) Centrifuge at 3000 rpm for 5 minutes | ||
| Line 64: | Line 66: | ||
:: 5) We labeled the purified stock as 7.15 | :: 5) We labeled the purified stock as 7.15 | ||
| - | : 3. Titer to determine phage concentration | + | : 3. Titer to determine phage concentration 1(7.15) |
:: 1) For each concentration of phage, specifically 10uL, 100uL, and 1mL, 1:100 dilution series were performed to generate 0, -2, -4, -6. 1:10 dilution series were then performed to generate -7, -8, and -9. | :: 1) For each concentration of phage, specifically 10uL, 100uL, and 1mL, 1:100 dilution series were performed to generate 0, -2, -4, -6. 1:10 dilution series were then performed to generate -7, -8, and -9. | ||
:: 2) For each dilution, a test tube was assigned to it. To each test tube, 0.75mL of BL21 liquid culture overnight was added and then transfected with 30uL of phage dilution. After 20 minutes of incubation, 7mL of x8 agar was added to each test tube and the content was then plated. | :: 2) For each dilution, a test tube was assigned to it. To each test tube, 0.75mL of BL21 liquid culture overnight was added and then transfected with 30uL of phage dilution. After 20 minutes of incubation, 7mL of x8 agar was added to each test tube and the content was then plated. | ||
| Line 74: | Line 76: | ||
:: 10uL phage: 4mL LB + 1mL E coli + 10μL 5.3 phage stock | :: 10uL phage: 4mL LB + 1mL E coli + 10μL 5.3 phage stock | ||
| - | : 5. Liquid culture purification (7.19) | + | : 5. Liquid culture purification 2 (7.19) |
| - | :: 1) For each infected test tubes (5uL | + | :: 1) For each infected test tubes (5uL and 10uL), put 1mL of liquid culture into 5 eppendorf tubes. (The 5th eppendorf tube for each culture only had about 0.75mL). |
| - | :: 2) Centrifuge at 3000 rpm for 5 minutes | + | :: 2) Centrifuge at 3000 rpm for 5 minutes. |
:: 3) Transfer supernatant into new eppendorf tubes. 5 eppendorf tubes for each phage concentration. | :: 3) Transfer supernatant into new eppendorf tubes. 5 eppendorf tubes for each phage concentration. | ||
| - | :: 4) Add 100 uL of chloroform to each of the new tubes and gently shake | + | :: 4) Add 100 uL of chloroform to each of the new tubes and gently shake. |
| - | :: 5) We labeled the purified stock as 7.19 | + | :: 5) We labeled the purified stock as 7.19. |
| - | : 6. Titer to determine phage concentration (7.19) | + | : 6. Titer to determine phage concentration 2 (7.19) |
:: 1) For each concentration of phage, specifically 5uL and 10uL, 1:100 dilution series were performed to generate 0, -2, -4, -6. 1:10 dilution series were then performed to generate -7, -8, and -9. | :: 1) For each concentration of phage, specifically 5uL and 10uL, 1:100 dilution series were performed to generate 0, -2, -4, -6. 1:10 dilution series were then performed to generate -7, -8, and -9. | ||
| - | :: 2) For each dilution, a test tube was assigned to it. To each test tube, 0.75mL of BL21 liquid culture overnight was added and then transfected with 30uL of phage dilution. After 20 minutes of incubation, 7mL of | + | :: 2) For each dilution, a test tube was assigned to it. To each test tube, 0.75mL of BL21 liquid culture overnight was added and then transfected with 30uL of phage dilution. After 20 minutes of incubation, 7mL of x1 agar was added to each test tube and the content was then plated. |
:: 3) Plates were incubated upside down for 24 hours. | :: 3) Plates were incubated upside down for 24 hours. | ||
'''V) Results''' | '''V) Results''' | ||
| - | |||
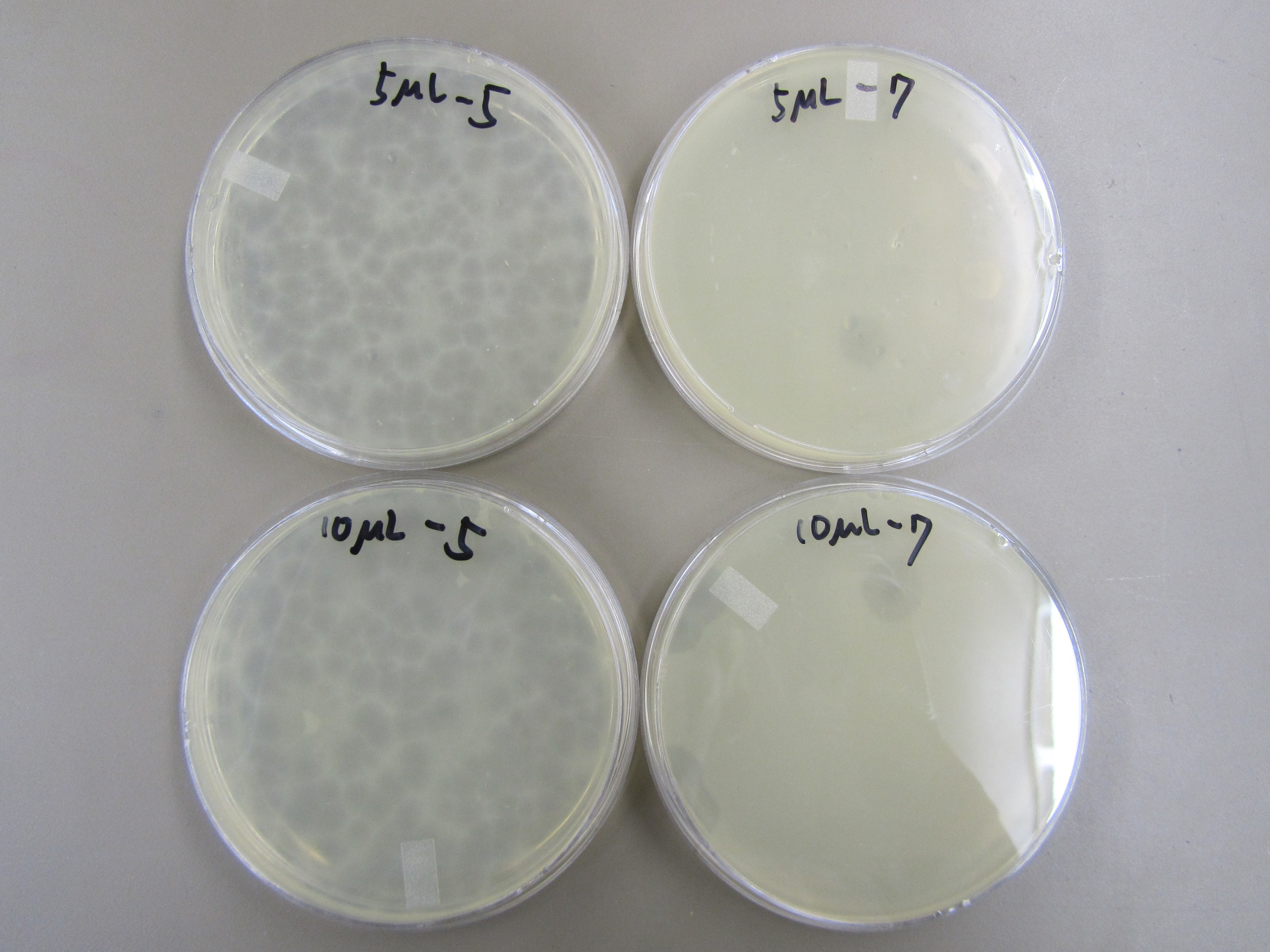
| - | : | + | : 3. Titer to determine phage concentration 1 |
| + | :: 10ug has many plaques on the -4 plate and 6 plaques on the -6 plate. | ||
| + | :: 100ug has many plaques on the -4 plate, none on the -6 plate, and 1 on the -7 plate. | ||
| + | :: 1mL has many plaques on the -4 plate, 4 on the -6 plate, and 2 on the -7 plate | ||
| + | :: We realized afterwards that using x8 top agar will not give us an accurate measure of phage concentration. From our results in [[Team:BYU Provo/Notebook/SmallPhage/Winterexp/Period3/Exp/4.26 T7 Phage Selection Test|4.26 T7 Phage Selection Test]], we know that the more concentrated the top agar is, the fewer the number of plaques will form on each plate. Thus for our next titer, we will use x1 agar. | ||
| + | |||
| + | : 4. Set up phage liquid culture - second round of propagation | ||
| + | :: The 10ug test tube was pretty clear, while the 5ug test tube was a little cloudy. The 0ug test tube was still very cloudy. | ||
| + | |||
| + | : 6. Titer to determine phage concentration 2 | ||
| + | :: Both 5ug and 10ug went down to -7. They appear to be at about the same concentration. | ||
| - | : | + | [[File:BYUSPSuProTiter.JPG|400px|center|link=]] |
| - | |||
'''VI) Conclusion/Next Step | '''VI) Conclusion/Next Step | ||
| + | :: After these two round of propagation, we are able to increase the titer of our phage stock to 10^(-9) PFU/mL. We will use this as the new stock for our future round of mutagenesis. Also, this experiment made us realize that phage titer will drop if it is stored inside the fridge for too long. This requires us to perform a titer periodically in the future to test for the viability of our phage stock. | ||
</font> | </font> | ||
Latest revision as of 15:35, 9 September 2013
| Small Phage July - August Notebook: Experiments
| ||
|
|
7.13 Phage Propagation Experiment
I) Purpose
II) Expected Outcome
III) Reagent Record
IV) Actual Procedure/Observations
V) Results
| |
 "
"