12/09/13
From 2013.igem.org
(Difference between revisions)
(→Double digestion of the isolated pGEM plasmids ligated with the TOD operon genes (X, F and TOBG)) |
(→Double digestion of the isolated pGEM plasmids ligated with the TOD operon genes (X, F and TOBG)) |
||
| Line 97: | Line 97: | ||
*To avoid the recircularization of the pSB1C3 backbone when ligating the genes with this plasmid, the double digestion was set up with the enzymes PstI and XbaI. | *To avoid the recircularization of the pSB1C3 backbone when ligating the genes with this plasmid, the double digestion was set up with the enzymes PstI and XbaI. | ||
*Another double digestion with EcoRI and SpeI was set up to test if the restrictions sites present in the TOD genes are not mutated. If the bands in the gel shows a linear band, it can be assumed that the enzymes cut the pGEM plasmids in its PstI and SpeI restriction sites. | *Another double digestion with EcoRI and SpeI was set up to test if the restrictions sites present in the TOD genes are not mutated. If the bands in the gel shows a linear band, it can be assumed that the enzymes cut the pGEM plasmids in its PstI and SpeI restriction sites. | ||
| + | |||
| + | ===Master Mix of the PstI/XbaI double digestion=== | ||
Revision as of 21:02, 13 September 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Contents |
Isolate plasmid from overnight culture
- Using Omega mini prep kit protocol
- Samples:
- TODF 1,2,3,
- TODX 1, 2, 3
- TOBG 1, 2, 3,
- RFP 1, 2, 3
| sample | Volume (ul) | Concentration (ng/ul) |
| TODF1 | 43 | 120 |
| TODF2 | 43 | 98.8 |
| TODX1 | 42 | 399.3 |
| TODX2 | 44 | 442.2 |
| TODX3 | 43 | 417.5 |
| TOBG1 | 43 | 262.4 |
| TOBG2 | 47 | 319.2 |
| TOBG3 | 41 | 338.4 |
| RFP1 | 41 | 77.7 |
| RFP2 | 41 | 32.7 |
| RFP3 | 42 | 59 |
Glycerol stocks
- Took 750ul from overnight culture and centrifuged at full speed for ~2 minutes
- Supernatant was removed
- Pellet resuspended in 375ul of HMFM
- Samples were then frozen at -80
Gel Purification of the pGEM-T pasmid double digestions ran in the 11/09/13
The ge was purified by using the The Zymoclean Gel DNA recovery Kit and its protocol was followed.
Nanodrop Results
| Sample | Volume | Concentration (ng/ul) | 280/260 | 260/230 |
| TODX | 48.5 | 7.1 | 1.83 | 0.72 |
| TODF | 45.5 | 21.7 | 1.89 | 1.19 |
| TOBG | 47.7 | 9 | 1.77 | 0.70 |
| pSB1C3 (RFP) | 46.2 | 7.3 | 1.81 | 0.16 |
Digestion of the samples purified with the enzyme PstI-HF
| DNA volume | Volume | Cutsmart buffer | Enzyme volume | dH2O added to total volume of 60 ul |
| TODX | 23 | 6 | 1 | 30 |
| TODF | 22.5 | 6 | 1 | 30.5 |
| TOBG | 22 | 6 | 1 | 31 |
| pSB1C3 (RFP) | 20.5 | 6 | 1 | 32.5 |
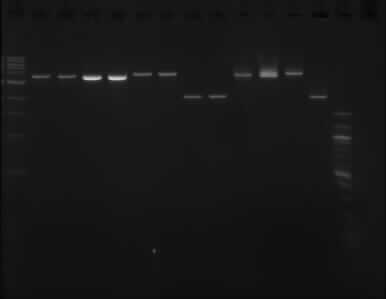
The picture above shows that the assumption of the PstI enzyme not working properly was wrong, as it shows the same result as the gel run on the 11/09/13.
Double digestion of the isolated pGEM plasmids ligated with the TOD operon genes (X, F and TOBG)
- As the digestions with the plasmids that were stored in the freezer did not work new digestins were set up with the isolated plasmids from the overnight culture.
- The pGEM-T plasmid contain the restrictions sites for PstI and SpeI, which are also present in the amplified TOD operon gene and the pSB1C3 plasmid.
- To avoid the recircularization of the pSB1C3 backbone when ligating the genes with this plasmid, the double digestion was set up with the enzymes PstI and XbaI.
- Another double digestion with EcoRI and SpeI was set up to test if the restrictions sites present in the TOD genes are not mutated. If the bands in the gel shows a linear band, it can be assumed that the enzymes cut the pGEM plasmids in its PstI and SpeI restriction sites.
 "
"