Team:BYU Provo/Notebook/CholeraDetection/Springexp/Period2/Dailylog
From 2013.igem.org
| (6 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{| width="100%" | {| width="100%" | ||
| - | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> '''Cholera Detection | + | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> |
| + | |||
| + | : '''Cholera Detection May - June Notebook: May 15 - May 26 Daily Log'''</font> | ||
<br> | <br> | ||
| Line 10: | Line 12: | ||
|- valign="top" | |- valign="top" | ||
| - | | style="width: | + | | style="width: 22%; background-color: transparent;"| |
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | : | + | <font size = "4"> |
| + | |||
| + | : <u> '''Cholera Detection''' </u> </font> | ||
: [[Team:BYU Provo/Notebook/Cholera_-_Detection/Winterexp|March-April]] | : [[Team:BYU Provo/Notebook/Cholera_-_Detection/Winterexp|March-April]] | ||
| Line 31: | Line 35: | ||
<font face="Calibri" size="3"> | <font face="Calibri" size="3"> | ||
| - | <font size="4"> ''' | + | <font size="4"> '''5/13/13''' </font> |
| - | + | Today we ran our digested CRO insert on the low melt gel, and under the UV light we saw the band we were looking for. Clarisse excised the band and we performed a ligation reaction according to the protocol and using the vector that we had already run on a low-melt gel last Thursday. After incubating for 30 minutes at room temperature, we performed a transformation into DH5alpha. The selectable marker of pLAT is ampicillin. | |
| - | + | We ran our low melt gel with our Cro insert. After 45 minutes, we put the gel under UV light and cut out the insert. We then performed a ligation of our vector and cro insert. After 30 min, we did a transformation and inserted our newly ligated plasmid into DH5alpha. | |
| - | KK, KP | + | KK, KP |
<br> | <br> | ||
| - | <font size="4"> ''' | + | <font size="4"> '''5/14/13''' </font> |
| + | |||
| + | (Thursday) When we dropped by today two of our plates (out of four) were contaminated. The two contaminated plates, we noticed, were the two plates that were old. The other two plates had a few colonies, but the was not a significantly greater number of colonies in our plasmid + insert + E.Coli plate than there were in our control plasmid + E.Coli plate. Kelton went ahead and streaked the few colonies we had to singles. We can PCR-verify if any of them have the plasmid. Meanwhile I am going to redo the transformation today. I will plate 4 plates: one 50 microL test, one 100 microL test, one 50 microL control, and one 100 microL control. | ||
| + | |||
| + | KK, KP | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <font size="4"> '''5/15/13''' </font> | ||
| + | |||
| + | KK Yesterday's transformation of pIG12+CRO insert into E.Coli yielded several colonies on both the plate inoculated with 50 microL and the plate inoculated 100 microL. We plated 8 separate single colonies and will wait until Friday, when we will plan to PCR verify which of the colonies has our vector and insert. Afterward, we discussed what we will do on Friday. We'll spend the hour preparing samples for PCR and a few hours later Clarisse is going to drop by to run the samples on a gel. | ||
| + | |||
| + | Also, I streaked lines and waves of V.cholerae today on a plate and for curiosity streaked E.Coli with pIG78 next to it in lines, and in waves intersecting the wavy cholera lines. We want to see if there are varying degrees of fluorescence. | ||
| + | |||
| + | KP 5/15/13 Today I re-streaked our ligation products. I re-streaked the control (pLat vector only) and the test (pLat vector and cro insert.) The hope, is that we can get individual colonies from the plates, so that we can sequence our plasmid to see if the vector took up the insert and if we have what we think that we do. We we will need primers IG57 and IG58 for our sequencing of this plasmid on Friday. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <font size="4"> '''5/17/13''' </font> | ||
| + | |||
| + | KK All our singles grew up well, so today we set up 10 PCR reactions + our control to test which, if any, had our vector + CRO insert construct. We also checked the plate that we streaked with Cholera and E.Coli with pIG78 next to one another. We wanted to see if the E.Coli would fluoresce at all in the presence of cholera. Under a UV light we didn't observe any noticeable difference between the streaks of cholera and the streaks of E.Coli | ||
| + | |||
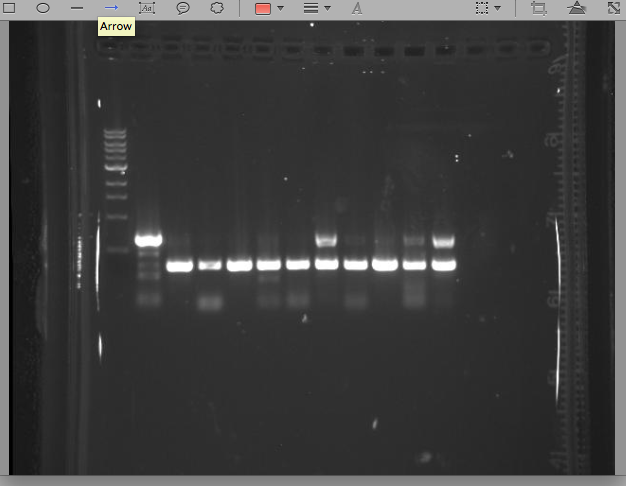
| + | Ran PCR with 10 singles we got from our ligations with pIG12/13 + Cro insert. See gel below. The bottom band in the ladder in the first lane runs at 500bps. Lanes 2-11 are the 10 strains we tested and the last lane is our control. What we get from this gel is a little unsure. | ||
| + | |||
| + | |||
| + | [[File:screen.jpg|250px|center]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <font size="4"> '''5/20/13''' </font> | ||
| + | |||
| + | KK Clarice left for Russia on Sunday, so she'll be gone for perhaps a month working at her internship. At the beginning of class we talked about possible community projects. The first project we may want to do is publish a children's book. The other two projects that received votes were: creating a board game, and sponsoring a fun run to raise money for Haiti. I called my friend Redge Ballard, an animation major, to ask if he would be interested in designing the book. | ||
| + | We PCR verified our 10 colonies and only colony A had the CRO insert we were looking for. We set up overnights of colony A to do a plasmid prep and then sequence, and we also set up overnights of our three lambda strains to plate tomorrow as a lawn. Once we do that, we hope to be induce lambda to go lytic by plating our E.Coli with CRO. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <font size="4"> '''5/22/13''' </font> | ||
| + | |||
| + | Yesterday I came in and did a plasmid prep of colonies A and W, which looked promising for having CRO+pIG12 on our PCR readout. Today we confirmed on the spectrophotometer the concentrations of the plasmids. The plasmid from A was present at a concentration of 158 ng/microL, and that of W had a concentration of 62 ng/microL. Yesterday we also plated our three strains of lambda-infected E.Coli, TT9901, TT9907, and TT23281, so today we were ready to trasform the plasmid into these three plasmids using electrophoration. Dr. Grose showed us how to use the electrophorator. We shocked each strain with plasmid from colony A and then a control plasmid separately, for a total of 6 electrophorations. After letting the cells recover for 30 minutes at 37 degrees celsius, we plated about 1 mL of each on LB/Amp plates. Tomorrow, hopefully, we will see a few colonies indicating that they took up our plasmid! Also, we submitted plasmids from A and W for sequencing. | ||
| + | |||
| + | KK | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <font size="4"> '''5/24/13''' </font> | ||
| + | |||
| + | KK, KP Our electrophoration worked on 2 of the 3 strains - on TT9901, and TT9907, but not on TT25281. Yesterday we went ahead and set up overnights to plate the two successful transformed strains on Amp/Arabinose, but our Amp/Arabinose plates are no longer good, so we need to make more. We redid the electroporation today so that hopefully by next wednesday all three strains should be ready and we should have plates. We'll be testing to see if TT9901, TT9907, and TT25281 with Lambda's CRO gene cloned into them can induce the integrated prophage out of lysogeny and into the lytic cycle. | ||
<br> | <br> | ||
{{TeamBYUProvoFooter}} | {{TeamBYUProvoFooter}} | ||
Latest revision as of 23:10, 27 September 2013
| ||
|
|
5/13/13 Today we ran our digested CRO insert on the low melt gel, and under the UV light we saw the band we were looking for. Clarisse excised the band and we performed a ligation reaction according to the protocol and using the vector that we had already run on a low-melt gel last Thursday. After incubating for 30 minutes at room temperature, we performed a transformation into DH5alpha. The selectable marker of pLAT is ampicillin. We ran our low melt gel with our Cro insert. After 45 minutes, we put the gel under UV light and cut out the insert. We then performed a ligation of our vector and cro insert. After 30 min, we did a transformation and inserted our newly ligated plasmid into DH5alpha. KK, KP
5/14/13 (Thursday) When we dropped by today two of our plates (out of four) were contaminated. The two contaminated plates, we noticed, were the two plates that were old. The other two plates had a few colonies, but the was not a significantly greater number of colonies in our plasmid + insert + E.Coli plate than there were in our control plasmid + E.Coli plate. Kelton went ahead and streaked the few colonies we had to singles. We can PCR-verify if any of them have the plasmid. Meanwhile I am going to redo the transformation today. I will plate 4 plates: one 50 microL test, one 100 microL test, one 50 microL control, and one 100 microL control. KK, KP
5/15/13 KK Yesterday's transformation of pIG12+CRO insert into E.Coli yielded several colonies on both the plate inoculated with 50 microL and the plate inoculated 100 microL. We plated 8 separate single colonies and will wait until Friday, when we will plan to PCR verify which of the colonies has our vector and insert. Afterward, we discussed what we will do on Friday. We'll spend the hour preparing samples for PCR and a few hours later Clarisse is going to drop by to run the samples on a gel. Also, I streaked lines and waves of V.cholerae today on a plate and for curiosity streaked E.Coli with pIG78 next to it in lines, and in waves intersecting the wavy cholera lines. We want to see if there are varying degrees of fluorescence. KP 5/15/13 Today I re-streaked our ligation products. I re-streaked the control (pLat vector only) and the test (pLat vector and cro insert.) The hope, is that we can get individual colonies from the plates, so that we can sequence our plasmid to see if the vector took up the insert and if we have what we think that we do. We we will need primers IG57 and IG58 for our sequencing of this plasmid on Friday.
5/17/13 KK All our singles grew up well, so today we set up 10 PCR reactions + our control to test which, if any, had our vector + CRO insert construct. We also checked the plate that we streaked with Cholera and E.Coli with pIG78 next to one another. We wanted to see if the E.Coli would fluoresce at all in the presence of cholera. Under a UV light we didn't observe any noticeable difference between the streaks of cholera and the streaks of E.Coli Ran PCR with 10 singles we got from our ligations with pIG12/13 + Cro insert. See gel below. The bottom band in the ladder in the first lane runs at 500bps. Lanes 2-11 are the 10 strains we tested and the last lane is our control. What we get from this gel is a little unsure.
5/20/13 KK Clarice left for Russia on Sunday, so she'll be gone for perhaps a month working at her internship. At the beginning of class we talked about possible community projects. The first project we may want to do is publish a children's book. The other two projects that received votes were: creating a board game, and sponsoring a fun run to raise money for Haiti. I called my friend Redge Ballard, an animation major, to ask if he would be interested in designing the book. We PCR verified our 10 colonies and only colony A had the CRO insert we were looking for. We set up overnights of colony A to do a plasmid prep and then sequence, and we also set up overnights of our three lambda strains to plate tomorrow as a lawn. Once we do that, we hope to be induce lambda to go lytic by plating our E.Coli with CRO.
5/22/13 Yesterday I came in and did a plasmid prep of colonies A and W, which looked promising for having CRO+pIG12 on our PCR readout. Today we confirmed on the spectrophotometer the concentrations of the plasmids. The plasmid from A was present at a concentration of 158 ng/microL, and that of W had a concentration of 62 ng/microL. Yesterday we also plated our three strains of lambda-infected E.Coli, TT9901, TT9907, and TT23281, so today we were ready to trasform the plasmid into these three plasmids using electrophoration. Dr. Grose showed us how to use the electrophorator. We shocked each strain with plasmid from colony A and then a control plasmid separately, for a total of 6 electrophorations. After letting the cells recover for 30 minutes at 37 degrees celsius, we plated about 1 mL of each on LB/Amp plates. Tomorrow, hopefully, we will see a few colonies indicating that they took up our plasmid! Also, we submitted plasmids from A and W for sequencing. KK
5/24/13 KK, KP Our electrophoration worked on 2 of the 3 strains - on TT9901, and TT9907, but not on TT25281. Yesterday we went ahead and set up overnights to plate the two successful transformed strains on Amp/Arabinose, but our Amp/Arabinose plates are no longer good, so we need to make more. We redid the electroporation today so that hopefully by next wednesday all three strains should be ready and we should have plates. We'll be testing to see if TT9901, TT9907, and TT25281 with Lambda's CRO gene cloned into them can induce the integrated prophage out of lysogeny and into the lytic cycle.
|
|
 "
"