Team:UCLA/Notebook/Library
From 2013.igem.org
(Difference between revisions)
Michaelc1618 (Talk | contribs) |
|||
| (One intermediate revision not shown) | |||
| Line 38: | Line 38: | ||
<li><a href="/Team:UCLA/Modeling">MODELING</a></li> | <li><a href="/Team:UCLA/Modeling">MODELING</a></li> | ||
<li><a href="/Team:UCLA/HumanPractices">HUMAN PRACTICES</a></li> | <li><a href="/Team:UCLA/HumanPractices">HUMAN PRACTICES</a></li> | ||
| - | <li><div id="spec"><a href="/Team:UCLA/Notebook"><font color="black">NOTEBOOK</font></a></div></li> | + | <li><div id="spec"><a href="/Team:UCLA/Notebook/Biobrick"><font color="black">NOTEBOOK</font></a></div></li> |
<li><a href="/Team:UCLA/Safety">SAFETY</a></li> | <li><a href="/Team:UCLA/Safety">SAFETY</a></li> | ||
<li><a href="/Team:UCLA/Attributions">ATTRIBUTIONS</a></li> | <li><a href="/Team:UCLA/Attributions">ATTRIBUTIONS</a></li> | ||
| Line 54: | Line 54: | ||
<html> | <html> | ||
<ul id="subnav"> | <ul id="subnav"> | ||
| - | <li style="margin-left: 12px;" | + | |
| - | + | <li style="margin-left: 12px;"><a href="https://2013.igem.org/Team:UCLA/Notebook/Biobrick">Mtd Biobrick Construction</a></li> | |
<li id = "current"><a href="/Team:UCLA/Notebook/Library">mRNA Display Library Generation</a></li> | <li id = "current"><a href="/Team:UCLA/Notebook/Library">mRNA Display Library Generation</a></li> | ||
<li><a href="/Team:UCLA/Notebook/mRNAdisplay">mRNA Display</a></li> | <li><a href="/Team:UCLA/Notebook/mRNAdisplay">mRNA Display</a></li> | ||
Latest revision as of 23:29, 27 September 2013
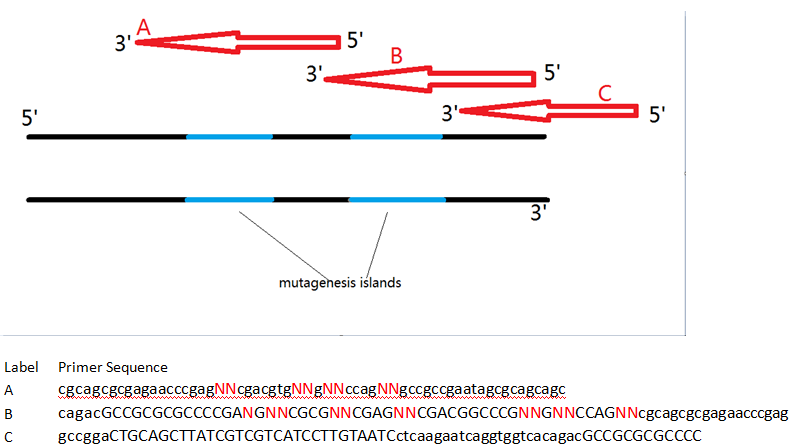
Generating the Mtd Library
Using PCR, we made several modifications to the mtd gene in order to generate our diverse library of mtd variants in a mRNA display-compatible format. Two sequential PCRs were used to generate the library. Primers and protocols are listed below.
| Primer Number | Primer Sequence |
|---|---|
| P10 Forward Primer | TATTGTAATACGACTCACTATAGGGCATTAGGAGGccaaCATTGCCACCatgagtaccgcagtccagttccg |
| P11 Primer C | gccggaCTGCAGCTTATCGTCGTCATCCTTGTAATCctcaagaatcaggtggtcacagacGCCGCGCGCCCC |
| P12 Primer B | cagacGCCGCGCGCCCCGANGNNCGCGNNCGAGNNCGACGGCCCGNNGNNCCAGNNcgcagcgcgagaacccgag |
| P13 Primer A | cgcagcgcgagaacccgagNNcgacgtgNNgNNccagNNgccgccgaatagcgcagcagc |
| P14 Splint | TTTTTTTTTTTTgccggaCTGCAG |
Generation of mtd library containing first mutagenic island
| Reagent | Volume (uL) |
|---|---|
| Water | 11.8 |
| Phusion Buffer HF | 4 |
| DMSO | 0.6 |
| dNTP (10 mM) | 0.4 |
| Bordetella Phage Genomic Template | 1 |
| P10 Forward Primer (10 uM) | 1 |
| P13 Primer A (10 uM) | 1 |
| Phusion DNA Polymerase | 0.2 |
| # Cycles | Temperature (°C) | Time |
|---|---|---|
| 1 | 98 | 0:30 |
| 30 | 98 65 72 | 0:10 0:20 0:25 |
| 1 | 72 | 8:00 |
| 1 | 4 | -- |
Gel extract and purify sample.
Generation of full mtd library containing second mutagenic island, FLAG tag, and splint attachment site
In this bridging PCR, a higher concentration of the outermost primer was used relative to the middle primer in order to maximize the chance that all products are full-length (both the middle and outermost primers bind). Equal concentrations of primers resulted in a broad ladder smear.
| Reagent | Volume (uL) |
|---|---|
| Water | 3.9 |
| KOD 2X Xtreme Buffer | 10 |
| dNTPs (10 mM) | 4 |
| P10 Forward Primer (10 uM) | 0.6 |
| P12 Primer B (10 uM) | 0.1 |
| P16 Primer A (10 uM) | 0.5 |
| mtd PCR Product | 0.5 (20 ng) |
| KOD Xtreme Hot Start Polymerase | 0.4 |
| # Cycles | Temperature (°C) | Time |
|---|---|---|
| 1 | 94 | 2:00 |
| 35 | 98 64 68 | 0:10 0:30 1:20 |
| 1 | 4 | -- |
 "
"