Results
A modified version of the green fluorescent protein (GFP) derived from ''Aequorea victoria'' with enhanced fluorescence properties. These enhanced fluorescent properties of the eGFP dramatically improved the spectral characteristics of GFP, resulting in increased fluorescence and photostability will increase even further the usefulness of the protein as reporter and for tracking of expression and subcellular localisation of proteins. Sequence was PCR amplified from pK7WGF2.0 described by Karimi et. al ("Gateway vectors for Agrobacterium-mediated plant transformation". Trends Plant Sci. 2002 May;7(5): 193-195).
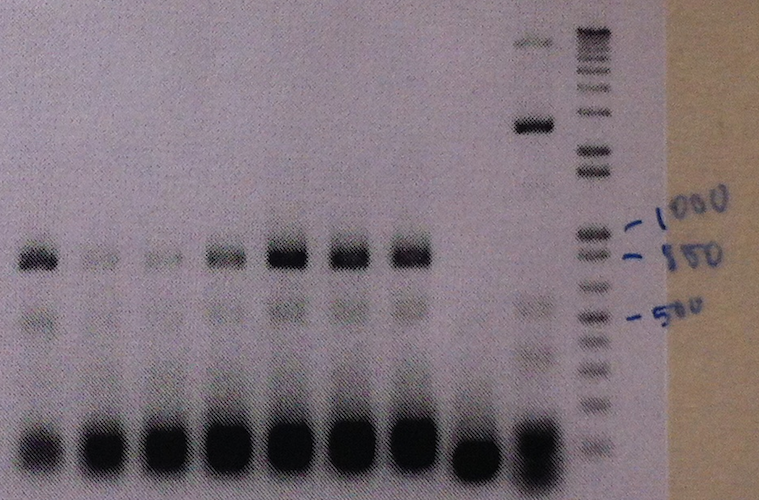
The part was cloned into the expression vector pJAM1786 via pDONR207 in ''E. coli'', by using the Gateway system by Invitrogen. Colony PCR and gel electrophoresis was performed to ensure the insert was in the vector (shown in the seven leftmost lanes of the figure below).
The restreaks of those seven colonies were inoculated into liquid culture and grown overnight alongside three E. coli cultures containing empty plasmid (pBBR1MCS-2). Fluorescence was measured in an opaque ELISA reader plate. The following dilutions were made: 1X, 2X, 5X, 10X and 50X. OD(600nm) was measured to ensure similar cell density. Excitation was carried out at 485 nm and emission was collected at 535 nm.
The mean values of the fluorescence can be found in the table below. Standard error for both groups (Total SE) was assessed using a Satterthwaite approximation.

(The raw data can be found here)
All dilution showed significantly more fluorescence than the control cultures except for the 50X dilution. The mean of the difference to the control cultures is shown as a function of dilution below.

A Western blot of the eGFP-pJAM1786 cultures was made with anti-GFP as primary antibodies. The eGFP-pJAM1786 samples give rise to band of the same size as a GFP positive control (see figure below). Unfortunately, the size standard was lost during the transfer to the membrane.

 "
"


