Team:Heidelberg/Templates/Del week15 overview
From 2013.igem.org
(Created page with "==Strategy== pFSN plasmid including Primers FS_01 to FS_3...") |
|||
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | |||
==Strategy== | ==Strategy== | ||
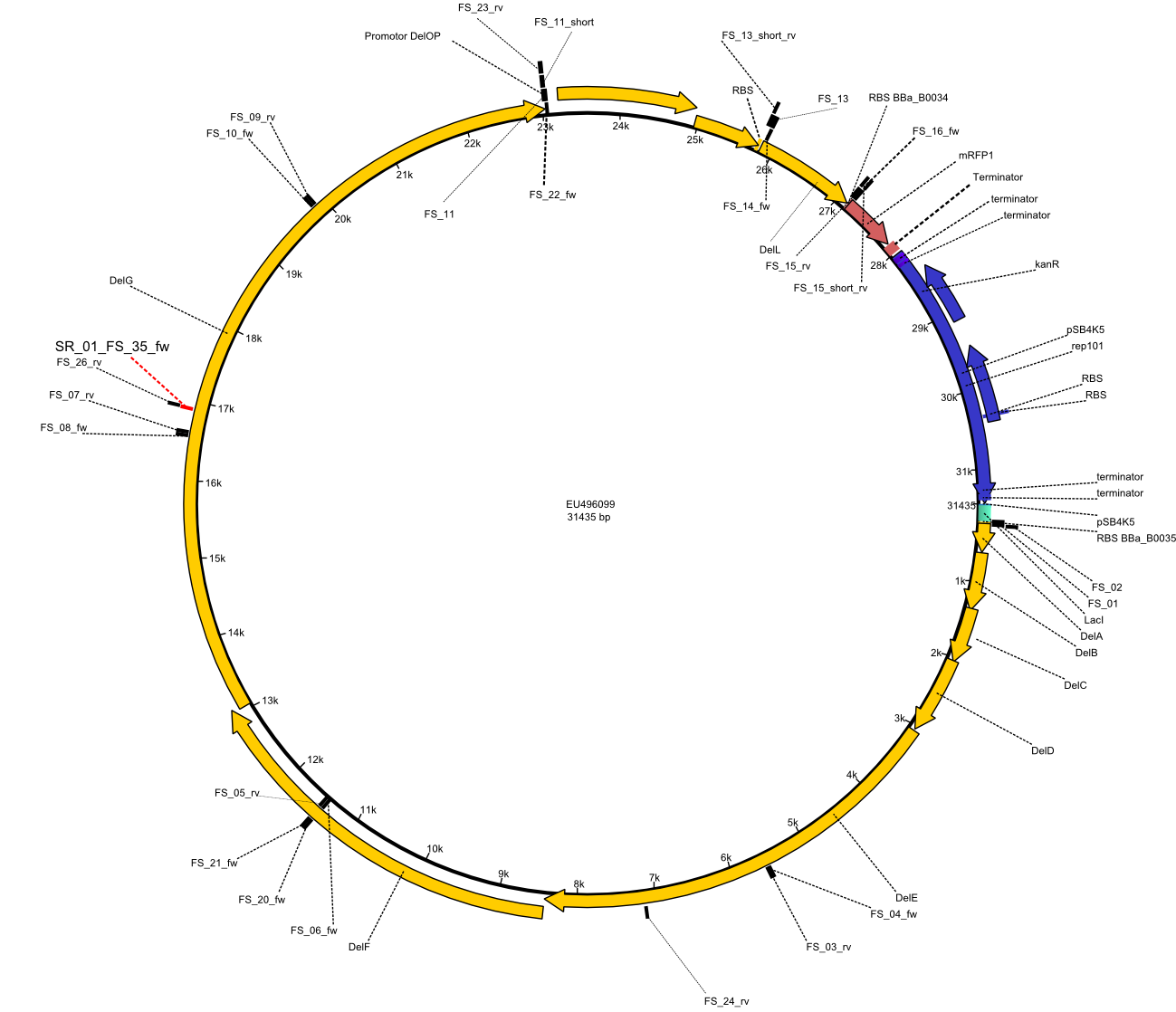
| - | + | [[File:Heidelberg_Strategie4.png | 300px|right|thumb| Vector map of [[:File:Heidelberg_Psb4k5+BBa J04450 DelRest.gb| pFSN plasmid]] including Primers FS_01 to FS_35/SR_01 used for assembly of the desired genes from the ''D. acidovorans'' genome and the partsregistry backbone pSB4K5 .]] | |
| - | + | ||
| - | [[File:Heidelberg_Strategie4.png | | + | |
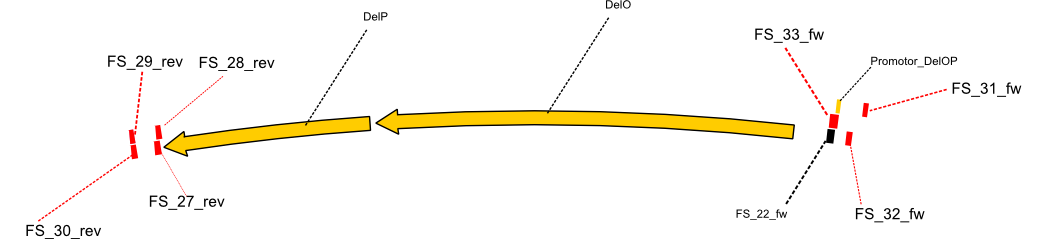
[[File:Heidelberg_Strategie5.png | 400px|right|thumb| Primers designed for a nested PCRs of the fragment DelO-P. The primers are shown on the Del-cluster as they are no more situated in DelO or DelP.]] | [[File:Heidelberg_Strategie5.png | 400px|right|thumb| Primers designed for a nested PCRs of the fragment DelO-P. The primers are shown on the Del-cluster as they are no more situated in DelO or DelP.]] | ||
As already indicated last week, we decided to order another primer for the sequence region of DelF-G, beeing the primer FS_35_SR_01. This primer shows significantly better secondary structures and therefore annealing should be better. | As already indicated last week, we decided to order another primer for the sequence region of DelF-G, beeing the primer FS_35_SR_01. This primer shows significantly better secondary structures and therefore annealing should be better. | ||
| Line 9: | Line 8: | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| + | ====Primer pairs, corresponding sequences and usage==== | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 19: | Line 19: | ||
==Cultivation of ''D. acidovorans'' substrain SPH-1== | ==Cultivation of ''D. acidovorans'' substrain SPH-1== | ||
Single read sequencing of several PCR products obtained so far was carried out by GATC. We aligned the so obtained sequences to the SPH-1 reference genome. (6.8 Mbp, [http://www.ncbi.nlm.nih.gov/genome/659 NCBI Genome]). Our alignments revealed significant differences between the two ''D. acidovorans'' strains: DSM-39 which we used as PCR template and SPH-1 based on which we have designed all PCR primers. | Single read sequencing of several PCR products obtained so far was carried out by GATC. We aligned the so obtained sequences to the SPH-1 reference genome. (6.8 Mbp, [http://www.ncbi.nlm.nih.gov/genome/659 NCBI Genome]). Our alignments revealed significant differences between the two ''D. acidovorans'' strains: DSM-39 which we used as PCR template and SPH-1 based on which we have designed all PCR primers. | ||
| - | As result of this sequence analysis, the strain | + | As result of this sequence analysis, the strain <b><i>D. acidovorans</i> SPH-1</b> was ordered in order to have a suitable template for the primers we already designed and ordered so far. We hoped, that haveing the right strain at hand now, our PCRs should work better and we could hopefully proceed with the Gibson assembly soon. |
| - | Lyohilized ''D. Acidovorans'' SPH-1 cells obtained from DSMZ were reactivated with | + | Lyohilized ''D. Acidovorans'' SPH-1 cells obtained from DSMZ were reactivated with Reactivation_medium and cultivated in Acidovorax complex medium according to the recoomondations by the DSMZ. Subsequently, liquid culture were inocculated (for glycerol stocks) and bacteria were also spread onto ACM-agar plates. |
Then we repeated our PCRs either by doing colony-PCR from colonies on the ACM-agar plates or by using 1µL of glycerol stock as template. | Then we repeated our PCRs either by doing colony-PCR from colonies on the ACM-agar plates or by using 1µL of glycerol stock as template. | ||
==Amplification of Del Genes from ''D. acidovorans'' SPH-1 genome== | ==Amplification of Del Genes from ''D. acidovorans'' SPH-1 genome== | ||
| - | ===Goals=== | + | ====Goals==== |
Since we wanted to ensure that our amplicons fit the intended sequence and avoid any mutations resulting from primer missmatches all PCRs were repeated (even the previously successful ones), now using ''D. Acidovorans'' SPH-1 as template. Established PCR protocols will be repeated with the new strain as template and adopted if necessary. For the DelF-G fragment with which we have had problems before, we will try whether PCRs work better with the new template or the recently ordered primer FS_35/SR_01. The amplification of DelO-P will be repeated with the intially ordered Primers FS_22 and FS_13 to test whether the changed template also improves these PCRs. If it turns out, that primers are still not able to bind to the intended template, the abovementioned nested PCR strategy will be conducted as primers FS_27 to FS_33 also arrived this week. | Since we wanted to ensure that our amplicons fit the intended sequence and avoid any mutations resulting from primer missmatches all PCRs were repeated (even the previously successful ones), now using ''D. Acidovorans'' SPH-1 as template. Established PCR protocols will be repeated with the new strain as template and adopted if necessary. For the DelF-G fragment with which we have had problems before, we will try whether PCRs work better with the new template or the recently ordered primer FS_35/SR_01. The amplification of DelO-P will be repeated with the intially ordered Primers FS_22 and FS_13 to test whether the changed template also improves these PCRs. If it turns out, that primers are still not able to bind to the intended template, the abovementioned nested PCR strategy will be conducted as primers FS_27 to FS_33 also arrived this week. | ||
| - | + | ====Results==== | |
{| class="wikitable" | {| class="wikitable" | ||
| Line 73: | Line 73: | ||
| Primer FS_22 and FS_29 || [[File:Heidelberg_Red x.svg.png|20px|center]] | | Primer FS_22 and FS_29 || [[File:Heidelberg_Red x.svg.png|20px|center]] | ||
|- | |- | ||
| - | + | <br/> | |
| - | + | <br/> | |
| DelL || Primer FS_14 and FS_15 || [[File:Heidelberg_Yes check.svg.png|20px|center]] | | DelL || Primer FS_14 and FS_15 || [[File:Heidelberg_Yes check.svg.png|20px|center]] | ||
|- | |- | ||
| '''pSB4K5''' || pSB4K5 || Primer FS_01 and FS_16 || [[File:Heidelberg_Yes check.svg.png|20px|center]] | | '''pSB4K5''' || pSB4K5 || Primer FS_01 and FS_16 || [[File:Heidelberg_Yes check.svg.png|20px|center]] | ||
|} | |} | ||
| - | |||
<br/> | <br/> | ||
| - | + | <br/> | |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
Latest revision as of 23:25, 4 October 2013
Contents |
Strategy
As already indicated last week, we decided to order another primer for the sequence region of DelF-G, beeing the primer FS_35_SR_01. This primer shows significantly better secondary structures and therefore annealing should be better. Furthermore we ordered various primers suitable for a nested PCR of DelOP.
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_35: DelG_fw | 2013-08-05 | Amplification of DelG from Delftia acidovorans Gibson Primer | CATGCAGATCATCGTGGATGAATTC |
Cultivation of D. acidovorans substrain SPH-1
Single read sequencing of several PCR products obtained so far was carried out by GATC. We aligned the so obtained sequences to the SPH-1 reference genome. (6.8 Mbp, [http://www.ncbi.nlm.nih.gov/genome/659 NCBI Genome]). Our alignments revealed significant differences between the two D. acidovorans strains: DSM-39 which we used as PCR template and SPH-1 based on which we have designed all PCR primers. As result of this sequence analysis, the strain D. acidovorans SPH-1 was ordered in order to have a suitable template for the primers we already designed and ordered so far. We hoped, that haveing the right strain at hand now, our PCRs should work better and we could hopefully proceed with the Gibson assembly soon. Lyohilized D. Acidovorans SPH-1 cells obtained from DSMZ were reactivated with Reactivation_medium and cultivated in Acidovorax complex medium according to the recoomondations by the DSMZ. Subsequently, liquid culture were inocculated (for glycerol stocks) and bacteria were also spread onto ACM-agar plates. Then we repeated our PCRs either by doing colony-PCR from colonies on the ACM-agar plates or by using 1µL of glycerol stock as template.
Amplification of Del Genes from D. acidovorans SPH-1 genome
Goals
Since we wanted to ensure that our amplicons fit the intended sequence and avoid any mutations resulting from primer missmatches all PCRs were repeated (even the previously successful ones), now using D. Acidovorans SPH-1 as template. Established PCR protocols will be repeated with the new strain as template and adopted if necessary. For the DelF-G fragment with which we have had problems before, we will try whether PCRs work better with the new template or the recently ordered primer FS_35/SR_01. The amplification of DelO-P will be repeated with the intially ordered Primers FS_22 and FS_13 to test whether the changed template also improves these PCRs. If it turns out, that primers are still not able to bind to the intended template, the abovementioned nested PCR strategy will be conducted as primers FS_27 to FS_33 also arrived this week.
Results
| PCRs from D.acidovorans SPH-1 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-E | Primer FS_02 and FS_03 | ||
| DelA-F | Primer FS_02 and FS_05 | |||
| DelA-G | Primer FS_02 and FS_23 | |||
| Primer FS_02 and FS_11 | ||||
| Primer FS_02 and FS_26 | ||||
| DelF-G | Primer FS_21 and FS_26 | |||
| DelG | Primer FS_08 and FS_23 | |||
| Primer SR_01 and FS_23 | ||||
| DelO DelP DelL | DelO-P | Primer FS_22 and FS_13 | ||
| Primer FS_22 and FS_13_short | ||||
| Primer FS_22 and FS_27 | ||||
| Primer FS_33 and FS_13_short | ||||
| Primer FS_33 and FS_27 | ||||
| Primer FS_31 and FS_29 | ||||
| Primer FS_31 and FS_30 | ||||
| Primer FS_32 and FS_29 | ||||
| Primer FS_32 and FS_30 | ||||
| Primer FS_22 and FS_28 | ||||
| Primer FS_22 and FS_29 | ||||
| DelL | Primer FS_14 and FS_15 | |||
| pSB4K5 | pSB4K5 | Primer FS_01 and FS_16 | ||
| Test restriction digests of PCR amplified fragments | ||||
|---|---|---|---|---|
| Fragment | Primer | Digestion enzyme | Expected bands | |
| DelF-G | Primer FS_21 and FS_26 | ClaI | 2743, 1519, 1208 | |
| BglII | 5500 | |||
| DelG | Primer FS_08 and FS_23 | ClaI | 3415, 3115 | |
| DelO-P | Primer FS_22 and FS_13 | EcoRI-HF | 1883, 960 | |
 "
"