|
|
| (33 intermediate revisions not shown) |
| Line 15: |
Line 15: |
| | { | | { |
| | background-color: white; | | background-color: white; |
| - | background-image: url("/wiki/images/7/70/Hintergrund3.jpg"); | + | background-image: url("/wiki/images/5/5c/Hintergrund_Grün(ohne_Header%2C_Logo_und_Schlagschatten).jpg"); |
| | background-attachment: scroll; | | background-attachment: scroll; |
| | background-position: 50% 0; | | background-position: 50% 0; |
| Line 26: |
Line 26: |
| | { | | { |
| | background: white; | | background: white; |
| - | background-image: url('/wiki/images/2/2f/Header_(mit_Logo).jpg'); | + | background-image: url("/wiki/images/2/2f/Header_%28mit_Logo%29.jpg"); |
| | margin: 0 auto; | | margin: 0 auto; |
| | height:250px ; | | height:250px ; |
| Line 35: |
Line 35: |
| | background-repeat: no-repeat; | | background-repeat: no-repeat; |
| | background-size: 100% 100% ; | | background-size: 100% 100% ; |
| - | border-style: none; | + | border-style: none;;} |
| - | } | + | |
| | | | |
| | #p-logo { display:none;} | | #p-logo { display:none;} |
| Line 43: |
Line 42: |
| | h1 {font-size:1.8em; font-weight:400; margin:0 0 12px;} | | h1 {font-size:1.8em; font-weight:400; margin:0 0 12px;} |
| | | | |
| - | #taskbar | + | |
| | + | #mm_icon1 |
| | { | | { |
| - | position:absolute; | + | position: absolute; |
| - | top:10px; | + | top: 150px; |
| - | left:400px; | + | left: 30px; |
| - | z-index: 5;
| + | |
| | } | | } |
| | | | |
| - | </style>
| + | #mm_icon2 |
| - | <!-- Taskbar -->
| + | { |
| - | <div id="taskbar">
| + | position: absolute; |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt">
| + | top: 150px; |
| - | <img alt="Home_ausgewählt" src="/wiki/images/e/ef/Darmstadt_green_Home.jpg" width="70" height="30"></a>
| + | left: 350px; |
| | + | } |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/problem">
| + | #mm_icon3 |
| - | <img alt="Problem" src="/wiki/images/6/66/Darmstadt_green_Problem.jpg" width="100" height="30"></a>
| + | { |
| | + | position: absolute; |
| | + | top: 150px; |
| | + | left: 670px; |
| | + | } |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/solution">
| + | #abstracticon1 |
| - | <img alt="solution" src="/wiki/images/8/87/Darmstadt_green_Solution.jpg" width="100" height="30"></a>
| + | { |
| | + | position: absolute; |
| | + | top: 150px; |
| | + | left: 30px; |
| | + | background:white; |
| | + | filter:alpha(opacity=83); opacity:0.83; |
| | + | border:1px solid #aaaaaa; |
| | + | -moz-border-radius:15px; |
| | + | -khtml-border-radius:15px; |
| | + | border-radius:15px; |
| | + | } |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/result">
| + | #abstracticon2 |
| - | <img alt="result" src="/wiki/images/2/2c/Darmstadt_green_Result.jpg" width="80" height="30"></a>
| + | { |
| | + | position: absolute; |
| | + | top: 150px; |
| | + | left: 350px; |
| | + | background:white; |
| | + | filter:alpha(opacity=83); opacity:0.83; |
| | + | border:1px solid #aaaaaa; |
| | + | -moz-border-radius:15px; |
| | + | -khtml-border-radius:15px; |
| | + | border-radius:15px; |
| | + | } |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/safety">
| |
| - | <img alt="safety" src="/wiki/images/7/7a/Darmstadt_green_Safety.jpg" width="90" height="30"></a>
| |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/team">
| |
| - | <img alt="team" src="/wiki/images/a/a4/Darmstadt_green_Team.jpg" width="70" height="30"></a>
| |
| - | <br>
| |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/strategy">
| + | #abstracticon3 |
| - | <img alt="team" src="/wiki/images/a/a0/Darmstadt_green_Strategy.jpg" width="100" height="30"></a>
| + | { |
| | + | position: absolute; |
| | + | top: 150px; |
| | + | left: 670px; |
| | + | background:white; |
| | + | filter:alpha(opacity=83); opacity:0.83; |
| | + | border:1px solid #aaaaaa; |
| | + | -moz-border-radius:5px; |
| | + | -khtml-border-radius:15px; |
| | + | border-radius:15px; |
| | + | } |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/humanpractice">
| + | #taskbar |
| - | <img alt="team" src="/wiki/images/4/4f/Darmstadt_green_Human_Practice.jpg" width="150" height="30"></a>
| + | { |
| | + | position:absolute; |
| | + | top:10px; |
| | + | left:500px; |
| | + | z-index: 5; |
| | + | } |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/modelling">
| |
| - | <img alt="team" src="/wiki/images/0/06/Darmstadt_green_Modelling.jpg" width="110" height="30"></a>
| |
| | | | |
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook">
| + | ul { |
| - | <img alt="team" src="/wiki/images/f/f3/Darmstadt_green_Labbook.jpg" width="90" height="30"></a>
| + | list-style: none; |
| | + | |
| | + | } |
| | | | |
| | + | li:before { |
| | + | content: "• "; |
| | + | color: white; |
| | + | } |
| | | | |
| | | | |
| | | | |
| - | </div>
| + | dl.igemTUD2013gelpicture |
| | + | { |
| | + | border: 1px solid #000; |
| | + | background-color: #109f71; |
| | + | width: 110px; |
| | + | text-align: center; |
| | + | padding: 5px 5px 5px 5px; |
| | + | float: right; |
| | + | margin: 0 0 0 0; |
| | + | margin-left:15px |
| | + | } |
| | | | |
| - | </html>
| + | .igemTUD2013gelpicture dt |
| | + | { |
| | + | font-weight: bold; |
| | + | background-color: #131210; |
| | + | color: #959289; |
| | + | padding: 0 0; |
| | + | margin-bottom: 10px; |
| | + | } |
| | | | |
| - | <span style="color:white;">
| + | .igemTUD2013gelpicture dd img |
| | + | { |
| | + | border: 1px solid #000; |
| | + | width: 100px; |
| | + | height: 200px; |
| | + | } |
| | | | |
| - | {| style="color:green;background:none;" | + | .igemTUD2013gelpicture dd |
| - | !align="center"|[[Team:TU_Darmstadt/labbook|<span style="color:white;font-size:160%;"> Labbook |</span>]]
| + | { |
| - | !align="center"|[[Team:TU_Darmstadt/protocols|<span style="color:white;font-size:160%;"> Protocols |</span> ]]
| + | margin: 0; |
| - | !align="center"|[[Team:TU_Darmstadt/materials|<span style="color:white;font-size:160%;"> Materials |</span>]]
| + | padding: 5px 5px 5px 5px; |
| - | |}
| + | font-size: 100%; |
| | + | text-align: left; |
| | + | } |
| | | | |
| - | {| class="wikitable" <!-- generated with [[w:de:Wikipedia:Technik/Text/Basic/EXCEL-Tabellenumwandlung]] V1.9 -->
| |
| - | |- valign="top"
| |
| - | | align="right" width="30" height="15" | 13.08
| |
| - | |style="font-weight:bold" width="416" | gBLOCK assembly of CMK and TLO
| |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · PCR mixture
| |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · 1 µL of each gBLOCK
| |
| | | | |
| - | |-
| + | dl.igemTUD2013gelpicture2 |
| - | | align="right" height="15" valign="top" |
| + | { |
| - | | valign="top" | · CMK: B_01 – B_04
| + | border: 1px solid #000; |
| | + | background-color: #109f71; |
| | + | width: 210px; |
| | + | text-align: center; |
| | + | padding: 5px 5px 5px 5px; |
| | + | float: right; |
| | + | margin: 0 0 0 0; |
| | + | margin-left:15px |
| | + | } |
| | | | |
| - | |-
| + | .igemTUD2013gelpicture2 dt |
| - | | align="right" height="15" valign="top" |
| + | { |
| - | | valign="top" | · TLO: A_01 – A_06
| + | font-weight: bold; |
| | + | background-color: #131210; |
| | + | color: #959289; |
| | + | padding: 0 0; |
| | + | margin-bottom: 10px; |
| | + | } |
| | | | |
| - | |-
| + | .igemTUD2013gelpicture2 dd img |
| - | | align="right" height="15" valign="top" |
| + | { |
| - | | valign="top" | · 10 µL 5x Q5 Reaction Buffer
| + | border: 1px solid #000; |
| | + | width: 200px; |
| | + | height: 200px; |
| | + | } |
| | | | |
| - | |-
| + | .igemTUD2013gelpicture2 dd |
| - | | align="right" height="15" valign="top" |
| + | { |
| - | | valign="top" | · 2 µL dNTPs
| + | margin: 0; |
| | + | padding: 5px 5px 5px 5px; |
| | + | font-size: 100%; |
| | + | } |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · 1 µL Q5 Hot Start Polymerase
| |
| | | | |
| - | |-
| + | </style> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 10 µL 5x Q5 High GC Enhancer
| + | |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · 1 µL primer suffix-R (10 mM)
| |
| | | | |
| - | |-
| + | <center> |
| - | | align="right" height="15" valign="top" |
| + | <!-- central main menu --> |
| - | | valign="top" | · 1 µL primer prefix_R (10 mM)
| + | |
| | | | |
| - | |-
| + | <br> |
| - | | align="right" height="15" valign="top" |
| + | <br> |
| - | | align="right" valign="top" |
| + | |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · PCR program (40 cycles)
| |
| | | | |
| - | |-
| + | <!-- Taskbar --> |
| - | | align="right" height="15" valign="top" |
| + | <div id="taskbar"> |
| - | | valign="top" | · initial denaturation 94°C, 100s
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt"> |
| | + | <img alt="Home_ausgewählt" src="/wiki/images/e/ef/Darmstadt_green_Home.jpg" width="70" height="30"></a> |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/problem"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="Problem" src="/wiki/images/6/66/Darmstadt_green_Problem.jpg" width="100" height="30"></a> |
| - | | valign="top" | · denaturation 94°C, 55s
| + | |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · annealing 64°C, 55s
| |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/result"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="result" src="/wiki/images/2/2c/Darmstadt_green_Result.jpg" width="80" height="30"></a> |
| - | | valign="top" | · elongation 72°C, 120s
| + | |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/safety"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="safety" src="/wiki/images/7/7a/Darmstadt_green_Safety.jpg" width="90" height="30"></a> |
| - | | valign="top" | · final elongation 72°C, 300s
| + | |
| | | | |
| - | |- valign="top"
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/team"> |
| - | | align="right" height="15" |
| + | <img alt="team" src="/wiki/images/a/a4/Darmstadt_green_Team.jpg" width="70" height="30"></a> |
| - | | align="right" |
| + | <br> |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/strategy"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="team" src="/wiki/images/a/a0/Darmstadt_green_Strategy.jpg" width="100" height="30"></a> |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/humanpractice"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="team" src="/wiki/images/4/4f/Darmstadt_green_Human_Practice.jpg" width="150" height="30"></a> |
| - | | valign="top" | · gel displays bands of expected size à assembly was successful
| + | |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/modelling"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="team" src="/wiki/images/0/06/Darmstadt_green_Modelling.jpg" width="110" height="30"></a> |
| - | | align="right" valign="top" |
| + | |
| | | | |
| - | |- valign="top"
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook"> |
| - | | align="right" height="15" |
| + | <img alt="team" src="/wiki/images/4/4c/10._Labbook_(angewählt).jpg" width="90" height="30"></a> |
| - | |style="font-weight:bold" align="right" |
| + | |
| | | | |
| - | |- valign="top"
| |
| - | | align="right" height="15" | 13.08
| |
| - | |style="font-weight:bold" | Purification
| |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · using Wizard SV Gel and PCR Clean-Up System (Promega)
| |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · TLO c = 34,4 ng/µL
| |
| | | | |
| - | |-
| + | </div> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · CMK c = 35,6 ng/µL
| + | |
| | | | |
| - | |- valign="top"
| |
| - | | align="right" height="15" |
| |
| - | | align="right" |
| |
| | | | |
| - | |- valign="top"
| |
| - | | align="right" height="15" | 19.08
| |
| - | |style="font-weight:bold" | Overlap extension PCR
| |
| | | | |
| - | |-
| + | <br><br><br><br> |
| - | | align="right" height="15" valign="top" |
| + | <center> |
| - | | valign="top" | ·
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook"> |
| | + | <font size="8" color="#F0F8FF" face="Arial regular">Lab book |</font> |
| | + | </a> |
| | + | <a href="https://2013.igem.org/Team:TU_Darmstadt/materials"> |
| | + | <font size="8" color="#F0F8FF" face="Arial regular">Materials |</font> |
| | + | </a> |
| | + | <a href="https://2013.igem.org/Team:TU_Darmstadt/protocols"> |
| | + | <font size="8" color="#F0F8FF" face="Arial regular">Protocols</font> |
| | + | </a> |
| | + | </center> |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | reaction mixture (50 µL total volume)
| |
| | | | |
| - | |-
| + | <br><br> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 25 µL Q5 High Fidelity 2x Master Mix (NEB)
| + | |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · 1 µL template
| |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · 1 µL forward primer (10 mM)
| |
| | | | |
| - | |-
| + | <h2><font size="6" color="#F0F8FF" face="Arial regular">Detection construct</font></h2> <br> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL reverse primer (10 mM)
| + | |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · 22 µL nuclease-free H2O
| |
| | | | |
| - | |- valign="top"
| + | <font size="3" color="#F0F8FF" face="Arial regular"> |
| - | | align="right" height="15" |
| + | <p text-aligne:left style="margin-left:50px; margin-right:50px"> |
| - | | align="right" |
| + | |
| | | | |
| - | |-
| + | The assembly of our detection construct proved to be tricky. Not only the traditional cloning approach failed but also the assembly using synthesized gBlocks delivered no results. We finally turned to an In-Fusion cloning approach to assemble our construct. Visit our lab book to read more about our work flow. |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · template and primer specifications
| + | |
| | | | |
| - | |-
| + | </p></font> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD1: pSB1C3[BBa_K206000] + frag1_for + frag4_rev
| + | |
| | | | |
| - | |-
| + | <br><br> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · CMK: CMK gBLOCK assembly + frag2_for + frag2_rev
| + | |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook/DetectionConstruct"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="Labbookdetection" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> |
| - | | valign="top" | · terminator: pSB1AK3[BBa_B0014] + frag3_for + frag3_rev
| + | <font size="6" color="#F0F8FF" face="Arial regular">Visit our lab book for the detection construct</font><br> |
| | + | </a> |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · pBAD4: pSB1C3[BBa_K206000] + frag4_for + frag4_rev
| |
| | | | |
| - | |-
| + | <br><br> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · TLO: TLO gBLOCK assembly + frag5_for + frag5_rev
| + | |
| | | | |
| - | |-
| + | <h2><font size="6" color="#F0F8FF" face="Arial regular">Fluorescence proteins</font></h2> <br> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pSB1C3: pSB1C3[fsC] + vector_for + vector_rev
| + | |
| | | | |
| - | |- valign="top"
| + | <font size="3" color="#F0F8FF" face="Arial regular"> |
| - | | align="right" height="15" |
| + | <p text-aligne:left style="margin-left:50px; margin-right:50px"> |
| - | | align="right" |
| + | |
| | | | |
| - | |-
| + | The genes for our FRET pair, mKate and LssmOrange, were isolated by PCR, cloned into the biobrick vector and sequenced afterwards. The genes were also cloned into a commercially available expression vector in order to characterize the fluorescent proteins. Visit our lab book to read more about our work flow. |
| - | | align="right" height="15" valign="top" |
| + | <center> |
| - | | valign="top" | · PCR program (35 cycles)
| + | <body> |
| | + | <br> |
| | + | <br> |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · initial denaturation: 98°C, 60s
| |
| | | | |
| - | |-
| + | <img alt="mkate" src="/wiki/images/3/3a/Mkate-active_light1.png" width="50%" height="50%" align="right"> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation: 98°C, 45s
| + | |
| | | | |
| - | |-
| + | <img alt="mkateactive" src="/wiki/images/5/58/Mkate-active_surface_activesite4_light1.png" width="50%" height="50%" align="right"> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing: 60°C, 40s
| + | |
| | | | |
| - | |-
| + | <br> |
| - | | align="right" height="15" valign="top" |
| + | <br> |
| - | | valign="top" | · elongation: 72°C, 90s
| + | </p></font> |
| | | | |
| - | |-
| + | <br><br> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation: 72°C, 300s
| + | |
| | | | |
| - | |- valign="top"
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook/Biobricks"> |
| - | | align="right" height="15" |
| + | <img alt="Labbookbiobricks" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> |
| - | | align="right" |
| + | <font size="6" color="#F0F8FF" face="Arial regular">Visit our lab book for the fluorescence proteins</font><br> |
| | + | </a> |
| | + | <br> |
| | | | |
| - | |-
| + | <h2><font size="6" color="#F0F8FF" face="Arial regular">Safety</font></h2> <br> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · gel displays byproducts
| |
| | | | |
| - | |-
| + | <font size="3" color="#F0F8FF" face="Arial regular"> |
| - | | align="right" height="15" valign="top" |
| + | <p text-aligne:left style="margin-left:50px; margin-right:50px"> |
| - | | valign="top" | à subsequent purification and amplification of desired bands (framed)
| + | |
| | | | |
| - | |-
| + | The assembly of the blue-light sensitive pDawn construct from gBlocks failed. Although the construction of the pezT construct from gBlocks was successful, multiple transformations of pSB1C3-pezT failed. Visit our lab book to read more about our work flow. |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| | | | |
| - | |- valign="top"
| + | </p></font> |
| - | | align="right" height="15" | 19.08
| + | <br><br> |
| - | |style="font-weight:bold" | Purification
| + | |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/safety/Labjournal"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="Labbookbiobricks" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> |
| - | | valign="top" | · using Wizard SV Gel and PCR Clean-Up System (Promega)
| + | <font size="6" color="#F0F8FF" face="Arial regular">Visit our lab book for the safety construct</font><br /> |
| | + | </a> |
| | + | |
| | | | |
| - | |-
| + | <h2><font size="6" color="#F0F8FF" face="Arial regular">Cutinase</font></h2> <br> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD1 c = 10 ng/µL
| + | |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | valign="top" | · CMK c = 30 ng/µL
| |
| | | | |
| - | |-
| + | <font size="3" color="#F0F8FF" face="Arial regular"> |
| - | | align="right" height="15" valign="top" |
| + | <p text-aligne:left style="margin-left:50px; margin-right:50px"> |
| - | | valign="top" | · terminator c = 7 ng/µL
| + | |
| | | | |
| - | |-
| + | Here, we present the improvement approach of the <i>Fusarium Solani Cutinase</i> (FsC, PID: BBa K808025). |
| - | | align="right" height="15" valign="top" |
| + | The FsC was one of the PET cleavage enzymes from the iGEM Team TU-Darmstadt 2012. We improved this part |
| - | | valign="top" | · pBAD4 c = 11 ng/µL
| + | with a pelB leader and a reporter(mRFP1-His6) system with a TEV cleavage site. This construct is regulated by pBAD |
| | + | (PID: BBa I0500) and cloned into the pSB1C3 backbone. |
| | | | |
| - | |-
| + | </p></font> |
| - | | align="right" height="15" valign="top" |
| + | <center> |
| - | | valign="top" | · TLO c = 22 ng/µL
| + | <img alt="fsc" src="/wiki/images/0/0d/Fscpet2.png" width="50%" height="50%"> |
| | + | <br><br> |
| | | | |
| - | |-
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook/Cutinase"> |
| - | | align="right" height="15" valign="top" |
| + | <img alt="Labbookdetection" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> |
| - | | valign="top" | · pSB1C3 c = 16 ng/µL
| + | <font size="6" color="#F0F8FF" face="Arial regular">Visit our lab book for the cutinase</font><br /> |
| | + | </a> |
| | | | |
| - | |-
| |
| - | | align="right" height="15" valign="top" |
| |
| - | | align="right" valign="top" |
| |
| | | | |
| - | |- valign="top"
| + | </body> |
| - | | align="right" height="15" | 19.08
| + | |
| - | |style="font-weight:bold" | Amplification PCR of pBAD1, terminator and pBAD4
| + | |
| | | | |
| - | |-
| + | </html> |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · reaction mixture (25 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 12,5 µL Q5 High Fidelity 2x Master Mix (NEB)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL template
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL forward primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL reverse primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 9,5 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · template and primer specifications
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD1: pBAD1 (c = 10 ng/µL, 19.08) + frag1_for + frag4_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · terminator: terminator (c = 7 ng/µL, 19.08) + frag3_for + frag3_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD4: pBAD4 (c = 11 ng/µL, 19.08) + frag4_for + frag4_rev
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (35 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation: 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation: 98°C, 45s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing: 70°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation: 72°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation: 72°C, 300s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · analytical 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD1 shows single band of expected size, 70°C is the optimal annealing temperature
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · terminator and pBAD4 show no bands
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 19.08
| + | |
| - | |style="font-weight:bold" | Gradient PCR of the amplification of pBAD1, terminator and pBAD4
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · reaction mixture (25 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 12,5 µL Q5 High Fidelity 2x Master Mix (NEB)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 4 µL template
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL forward primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL reverse primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 6,5 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · template and primer specifications
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD1: pBAD1 (c = 10 ng/µL, 19.08) + frag1_for + frag4_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · terminator: terminator (c = 7 ng/µL, 19.08) + frag3_for + frag3_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD4: pBAD4 (c = 11 ng/µL, 19.08) + frag4_for + frag4_rev
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (35 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation 98°C, 45s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing 55°C/57°C/61°C/65°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation 72°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation 72°C, 300s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · analytical 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · optimal annealing temperature for terminator is 55°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · optimal annealing temperature for pBAD4 is 55°C
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 20.08
| + | |
| - | |style="font-weight:bold" | Purification of PCR batches pBAD1 (Amplification PCR, 19.08), terminator 55°C (Gradient PCR, 19.08) and pBAD4 55°C (Gradient PCR, 19.08)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · using Wizard SV Gel and PCR Clean-Up System (Promega)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD1 c = 45 ng/µL
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · terminator c = 44 ng/µL 260/280 = 1,67 260/230 = 0,73
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pBAD4 c = 57,7 ng/µL 260/280 = 1,81 260/230 = 0,67
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 21.08
| + | |
| - | |style="font-weight:bold" | Amplification PCR of CMK, TLO and pSB1C3
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 25 µL Q5 High Fidelity 2x Master Mix
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL template
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL forward primer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL reverse primer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 22 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · template and primer specifications
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · CMK: CMK (c = 30 ng/µL, 19.08) + frag2_for + frag2_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · TLO: TLO (c = 22 ng/µL, 19.08) + frag5_for + frag5_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pSB1C3: pSB1C3 (c = 16 ng/µL, 19.08) + vector_for + vector_rev
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (35 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation 98°C, 45s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing 55°C, 40s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation 72°C, 90s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation 72°C, 300s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · analytical 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · gel displays critical amount of byproducts
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 21.08
| + | |
| - | |style="font-weight:bold" | Gradient PCR of the amplification of CMK, TLO and pSB1C3
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · Amplification using Q5 Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 25 µL Q5 High Fidelity 2xMaster Mix
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL template
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL forward primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL reverse primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 22 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (35 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation 98°C, 45s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing 55°C/56,7°C/60,4°C/62,9°C/64,9°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation 72°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation 72°C, 300s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · Amplification using Pfu Polymerase (homehold)
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL template
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 10 µL 5x Phusion-Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL dNTPs
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 4 µL Pfu-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL MgCl2 (50 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL forward primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL reverse primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 26 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (30 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • annealing 55°C/56,4°C/59,3°C/62,1°C/65°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • elongation 72°C, 720s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • final elongation 72°C, 600s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · template and primer specifications
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · CMK: CMK (c = 30 ng/µL, 19.08) + frag2_for + frag2_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · TLO: TLO (c = 22 ng/µL, 19.08) + frag5_for + frag5_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pSB1C3: pSB1C3 (c = 16 ng/µL, 19.08) + vector_for + vector_rev
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · amplification with Pfu Polymerase yields lesser byproducts than amplification with Q5 polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · for all three templates higher annealing temperatures diminish the amount of by products
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 22.08
| + | |
| - | |style="font-weight:bold" | Purification of CMK, TLO and pSB1C3 from preparative gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · using Wizard SV Gel and PCR Clean-Up System (Promega)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · CMK c = 154,2 ng/µL 260/280 = 1,92 260/230 = 1,88
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · TLO c = 37,9 ng/µL 260/280 = 1,90 260/230 = 0,60
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pSB1C3 c = 120,8 ng/µL 260/280 = 1,89 260/230 = 1,64
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 23.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL TLO (c = 37,9 ng/µL, 22.08)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 10 µL 5x Phusion-Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL dNTPs
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 4 µL Pfu-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL MgCl2 (50 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL frag5_for (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 26 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (30 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • annealing 55°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • elongation 72°C, 300s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • final elongation 72°C, 600s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · gel displays no distinctive bands
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 24.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL TLO (c = 37,9 ng/µL, 22.08)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 10 µL 5x Phusion-Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL dNTPs
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 4 µL Pfu-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL MgCl2 (50 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL frag5_for (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 27 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (30 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • annealing 56°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • elongation 72°C, 720s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • final elongation 72°C, 600s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · gel displays no distinctive bands (just like on 23.08) à probably the template is poorly
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 24.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL TLO gBLOCK assembly (c = 34,4 ng/µL)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 10 µL 5x Phusion-Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL dNTPs
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 4 µL Pfu-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL MgCl2 (50 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL frag5_for (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 27 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (30 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • annealing 56°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • elongation 72°C, 720s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • final elongation 72°C, 600s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · gel displays no distinctive bands
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 25.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL TLO gBLOCK assembly (c = 34,4 ng/µL)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 10 µL 5x Phusion-Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL dNTPs
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 4 µL Pfu-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 2 µL MgCl2 (50 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL frag5_for (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 1 µL frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • 26 µL nuclease-free H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (30 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • annealing 55°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • elongation 72°C, 300s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • final elongation 72°C, 600s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · gel displays the desired band of ~2500 bp as well as byproducts
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 25.08
| + | |
| - | |style="font-weight:bold" | Purification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · using Wizard SV Gel and PCR Clean-Up System (Promega)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · TLO c = 19 ng/µL
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | |style="font-weight:bold" align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 25.08
| + | |
| - | |style="font-weight:bold" | InFusion
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · reaction mixture (71 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl pBAD1 (130 bp, c = 45 ng/µL, 45 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl pBAD4 (130 bp, c = 57,7 ng/µL, 58 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl terminator (100 bp, c = 44 ng/µL, 44 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 42µl TLO (2440 bp, c = 19 ng/µL, 780 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 4,5µl CMK (1850 bp, c = 154,2 ng/µL, 680 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 3,5µl pSB1C3 (2100 bp, c = 120,8 ng/µL, 360 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 18µl 5x InFusion Pfu MasterMix
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · treatment of the reaction mixture according to InFusion protocol
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · transformation into E.coli Top10 according to heat shock protocol
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · no colonies grew on LB-Cam plates, most likely the reaction volume was to high
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | |style="font-weight:bold" align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 26.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | ·
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | 1 µL TLO gBLOCK assembly (c = 34,4 ng/µL)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL frag5_for (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 10 µL 5x Phusion Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 27 µL nucleasefree H2O
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL dNTPs
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL MgCl2 (50 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 4 µL Pfu-Polymerase
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (30 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • annealing 55°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • elongation 72°C, 300s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • final elongation 72°C, 600s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · gel displays the desired band of ~2500 bp as well as byproducts
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 26.08
| + | |
| - | |style="font-weight:bold" | Purification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · using Wizard SV Gel and PCR Clean-Up System (Promega)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · TLO c = 19,7 ng/µL
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 26.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL TLO (c = 19,7 ng/µL)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL frag5_for (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 10 µL 5x Phusion Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 27 µL nucleasefree H2O
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL dNTPs
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL MgCl2 (50 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 4 µL Pfu-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (35 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • annealing 55°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • elongation 72°C, 300s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | • final elongation 72°C, 600s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · no distinctive bands
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 27.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL TLO (c = 19,7 ng/µL, 1:4 dilution, final amount = 5 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL frag5_for (1:10)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 10 µL 5x Phusion Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 25,5 µL nucleasefree H2O
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL dNTPs (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2,5 µL DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL MgCl2 (50 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 4 µL Pfu-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 µL BSA (10 mg/mL, final concentration = 0,4 µg/µL)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (35 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing 55°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation 72°C, 480s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation 72°C, 600s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · no distinctive bands (just like on 26.08)
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 28.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL TLO (c = 19,7 ng/µL, 1:20 dilution, final amount = 1 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2,5 µL primer frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2,5 µL primer frag5_for (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 19 µL nucleasefree H2O
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 25 µL 2x Q5 Mastermix (NEB)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (35 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing 66°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation 72°C, 90s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation 72°C, 120s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · no distinctive bands (just like on 26.08) à probably the template is poorly
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | |style="font-weight:bold" align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 28.08
| + | |
| - | |style="font-weight:bold" | Amplification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1 µL TLO gBLOCK assembly (c = 34,4 ng/µL, 1:30 dilution, final amount = 1 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | ·
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | 2,5 µL primer frag5_rev (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2,5 µL primer frag5_for (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 19 µL nucleasefree H2O
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 25 µL 2x Q5 Mastermix (NEB)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (35 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation 98°C, 60s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation 98°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing 66°C, 30s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation 72°C, 90s
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation 72°C, 120s
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · preparative 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · gel displays the desired band of ~2500 bp as well as byproducts
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | |style="font-weight:bold" align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 29.08
| + | |
| - | |style="font-weight:bold" | Purification of TLO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · using Wizard SV Gel and PCR Clean-Up System (Promega)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · TLO c = 241,7 ng/µL 260/280 = 1,93 260/230 = 2,02
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | |style="font-weight:bold" align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 31.08
| + | |
| - | |style="font-weight:bold" | InFusion
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · reaction mixture (20 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl pBAD1 (130 bp, c = 45 ng/µL, 45 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl pBAD4 (130 bp, c = 57,7 ng/µL, 58 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl terminator (100 bp, c = 44 ng/µL, 44 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 3,5µl TLO (2440 bp, c = 241,7 ng/µL, 780 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 4,5µl CMK (1850 bp, c = 154,2 ng/µL, 680 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 3,5µl pSB1C3 (2100 bp, c = 120,8 ng/µL, 360 ng)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 4µl 5x InFusion Pfu MasterMix
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1,5µl nucleasefree H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · treatment of the reaction mixture according to InFusion protocol
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · transformation into E.coli Top10 according to heat shock protocol
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2 colonies grew on LB-Cam plates, most likely the reaction volume was to high
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | |style="font-weight:bold" align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 2.09
| + | |
| - | |style="font-weight:bold" | Control of pSB1C3-CMK-TLO clone 1 and 2 using colony PCR and test restriction
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · Colony PCR
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl VR primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl VF2 primer (10 mM)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 5µl Colony LB Medium (Colony 1/Colony 2)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2µl MgCl2
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl dNTP Mix
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 5µl 10x Taq Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl Taq-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 33µl nucleasefree H2O
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR Programm
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation 300s 95°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation 30s 95°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing 30s 55°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation 150s 72°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation 300s 72°C
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · Purification using Pure Yield Plasmid Miniprep System (Promega)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · Colonie 1 = pSB1C3-TLO-CMK 1
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o c = 245,5 ng/ul
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o 260/280 = 1,87
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o 260/230 = 2,23
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · Colonie 2 = pSB1C3-TLO-CMK 2
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o c = 107,7 ng/ul
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o 260/280 = 1,95
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o 260/230 = 2,28
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · digestion with EcoRI and double digest with EcoRI and PstI
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · analytical 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · pattern is as expected
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · colony PCR product is off by 2000 bp
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · linearized plasmid is off by 1000 bp
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 3.09
| + | |
| - | |style="font-weight:bold" | Control of pSB1C3-CMK-TLO clone 1 and 2 using PCR
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR mixture (50 µL total volume)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl forward primer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl reverse primer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl template
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 2µl MgCl2
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl dNTP Mix
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl DMSO
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 5µl 10x Taq-Buffer
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 1µl Taq-Polymerase
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · 37µl H2O
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | align="right" valign="top" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · template and primer specifications
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · template = pSB1C3-TLO-CMK 1, c = 245,5 ng/µl, 1:10 dilution
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 1: frag1_for + frag4_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | PCR tube 2: frag2_for + frag2_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 3: frag3_for + frag3_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 4: frag4_for + frag4_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 5: frag5_for + frag5_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · template = pSB1C3-TLO-CMK 2, c = 107,7 ng/µl, 1:4 dilution
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 1: frag1_for + frag4_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 2: frag2_for + frag2_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 3: frag3_for + frag3_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 4: frag4_for + frag4_rev
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | o PCR tube 5: frag5_for + frag5_rev
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · PCR program (30 cycles)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · initial denaturation 300s 95°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · denaturation 30s 95°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · annealing 30s 55°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · elongation 150s 72°C
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · final elongation 300s 72°C
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · analytical 1% agarose gel
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · besides many byproducts all fragments
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | could be amplified from templates (framed bands)
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | à clones are most likely correct
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 3.09
| + | |
| - | |style="font-weight:bold" | Induction of pSB1C3-TLO-CMK clones with L-Arabinose
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · induction with L-arabinose (0,02% w/v) at OD600 = 0,6
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · induced clones showed no difference to native E.coli Top10 cells when analyzed with an fluorospcetrometer
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 3.09
| + | |
| - | |style="font-weight:bold" | Sequencing of pSB1C3-TLO-CMK clones
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · were not able to sequence the whole insert, as pimers did not anneal sufficiently
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · results indicate that all five fragments were successfully ligated in the correct order
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · results show that gBLOCK assembly of TLO and CMK did not work correctly
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 3.09
| + | |
| - | |style="font-weight:bold" | Induction of pSB1C3-TLO-CMK clones with L-Arabinose
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · induction with L-arabinose (0,02% w/v) at OD600 = 0,6
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · induced clones showed no difference to native E.coli Top10 cells when analyzed with an fluorospcetrometer
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | |
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" | 3.09
| + | |
| - | |style="font-weight:bold" | Sequencing of pSB1C3-TLO-CMK clones
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · were not able to sequence the whole insert, as pimers did not anneal sufficiently
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · results indicate that all five fragments were successfully ligated in the correct order
| + | |
| - | | + | |
| - | |-
| + | |
| - | | align="right" height="15" valign="top" |
| + | |
| - | | valign="top" | · results show that gBLOCK assembly of TLO and CMK did not work correctly
| + | |
| - | | + | |
| - | |- valign="top"
| + | |
| - | | align="right" height="15" |
| + | |
| - | | align="right" |
| + | |
| - | | + | |
| - | |}
| + | |
 Visit our lab book for the detection construct
Visit our lab book for the detection construct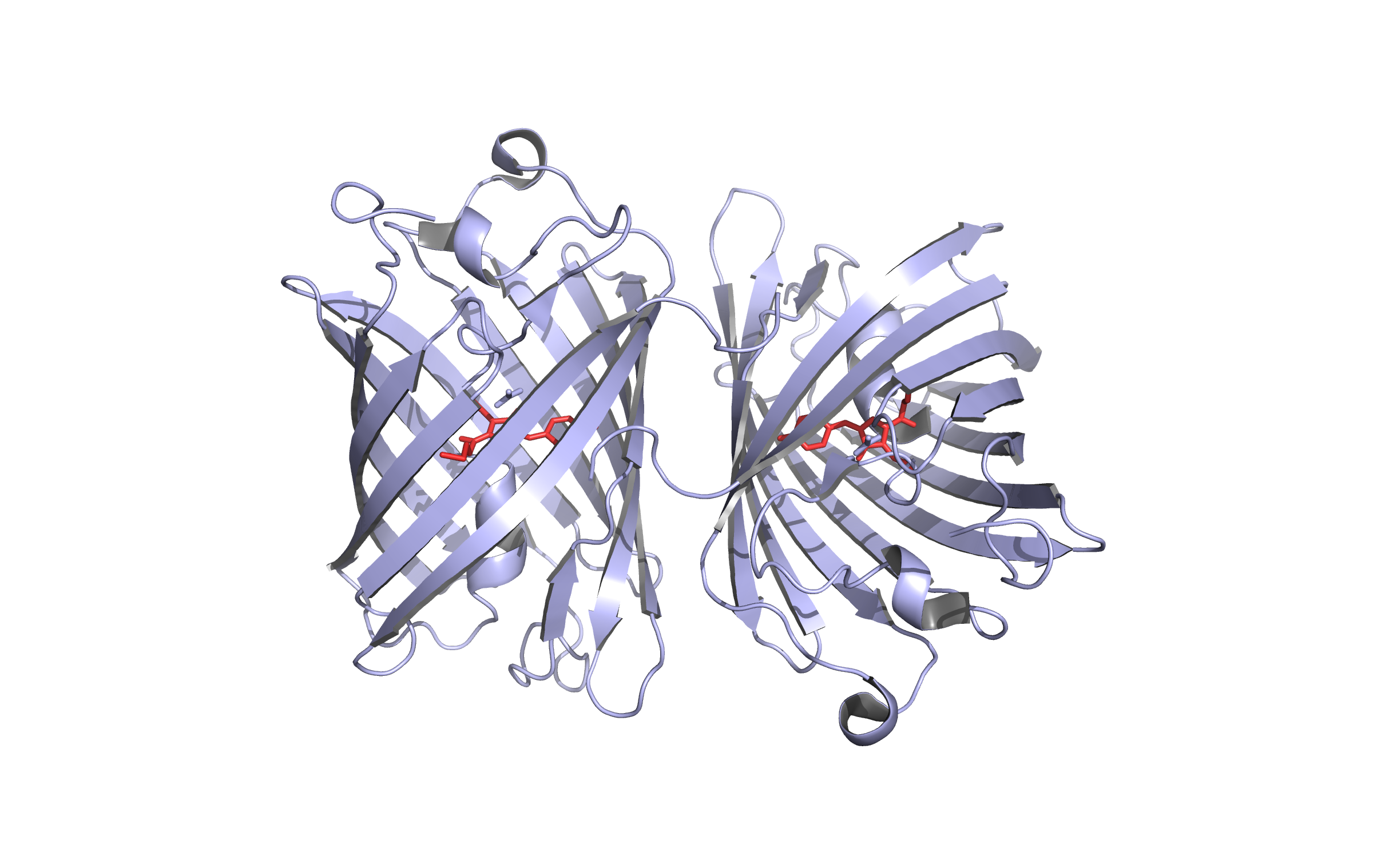
 Visit our lab book for the fluorescence proteins
Visit our lab book for the fluorescence proteins Visit our lab book for the safety construct
Visit our lab book for the safety construct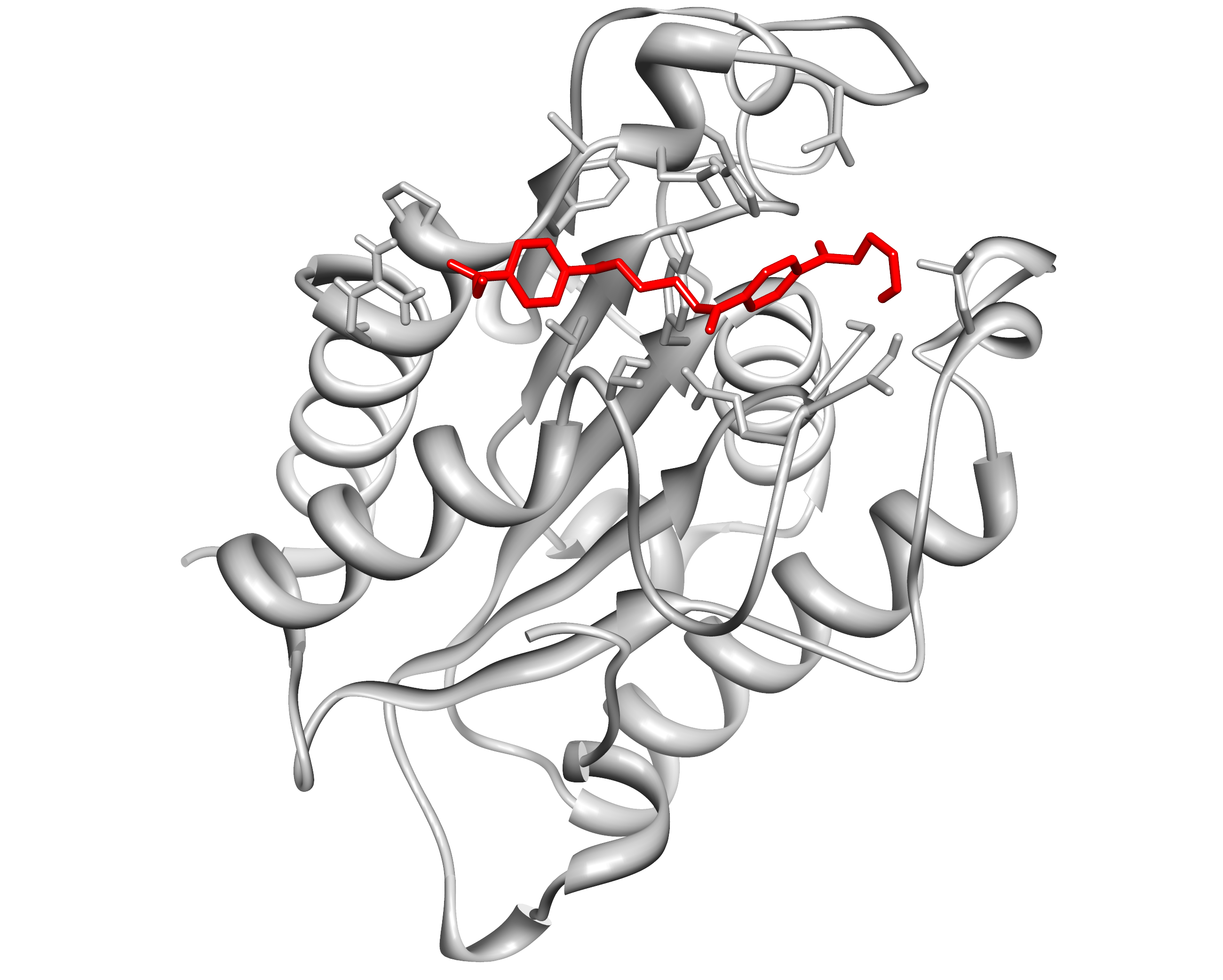
 Visit our lab book for the cutinase
Visit our lab book for the cutinase "
"