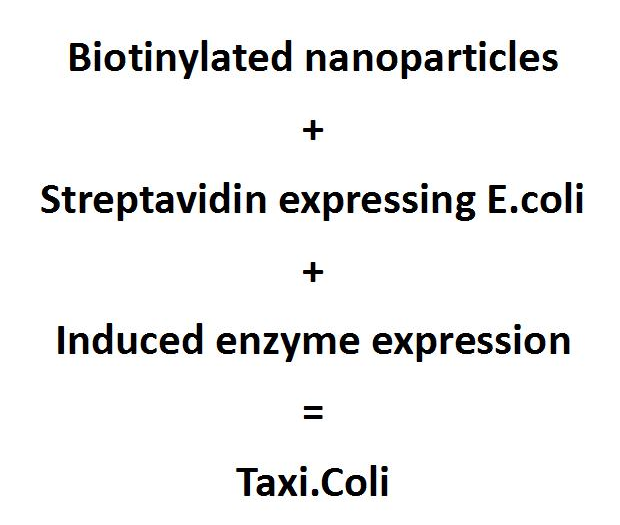|
|
| (25 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{Template:EPFL2013Header}} | | {{Template:EPFL2013Header}} |
| | | | |
| - | [[File:Team-EPF-Lausanne Overview-schema.png|thumb|300px |Figure 1: Assembling basic parts to make a smart drug delivery system: Taxi.Coli]] | + | [[File:Team-EPF-Lausanne Overview-schema.png|thumb|300px |Figure 1: Assembling the different modules to make a smart drug delivery system: Taxi.Coli]] |
| - | Nowadays when people get sick they ingest a pill or get an intravenous injection to combat the pathogens. The drug gets distributed throughout the entire body of the patient, and can cause severe side effects. The idea of targeted drug delivery is to increase the concentration of the drug at the target site while keeping it to a minimum in the rest of the organism. Such specific drug delivery can be used to treat various diseases, especially tumors, whether benign or malignant, or ulcers. In theory, the side effects of the drugs would be less severe. Especially spikes of drugs in the blood system would be avoided in treatments like chemotherapy. Furthermore, the amount of drugs that have to be ingested could be reduced. One big drawback of targeted drug delivery is that it doesn’t exist for many drugs, it needs to be developed for each drug separately and that is obviously quite expensive. <BR><BR><BR><BR><BR><BR>
| + | Smart drug delivery systems administer drugs, where they are needed, when they are needed and at the right concentration. |
| | + | Thus diminish the side effects and the amount of drugs needed. It could eventually be used to treat a wide range of diseases, but especially tumors. The idea to target the drug delivery is to increase the concentration of the drug at the target site while keeping it to a minimum in the rest of the organism. One big drawback of smart drug delivery is that it doesn't exist for many drugs, it needs to be developed for each drug separately and that is quite expensive. |
| | + | |
| | + | <BR><BR><BR> |
| | | | |
| | ==Introduction== | | ==Introduction== |
| - | Our team aims to create a highly adaptable smart drug delivery system, which could be used for several applications and alternatives in disease treatment. The design makes it so flexible that it could even be possible to adapt it to the patient’s individual needs. By being highly adaptive, the system we propose would also reduce the cost of targeted drug delivery because, instead of creating a new system for each drug and each patient, one could simply use the basic part that we made, modify them slightly and assemble them. This sort of "Parts recycling" would not only be economical but also ecological. | + | Our team aims to create a highly adaptable smart drug delivery system, which could be used for several applications and alternatives in disease treatment. The design makes it so flexible that it could even be possible to adapt it to the patient’s individual needs. By being highly adaptive, the system we propose would also reduce the cost of targeted drug delivery because, instead of creating a new system for each drug and each patient, one could simply use the basic part that we made, modify them slightly and assemble them. |
| | | | |
| | ===A Proof of Principle=== | | ===A Proof of Principle=== |
| | Our effort is limited to a proof of principle. We used E.coli as model organism. Using the principles of synthetic biology we aimed to engineer an E.coli Bacterium that expresses streptavidin on its surface and contains a pH sensing promoter that controls the expression of gelatinases. Those bacteria would be able to bind biotinylated nanoparticles. | | Our effort is limited to a proof of principle. We used E.coli as model organism. Using the principles of synthetic biology we aimed to engineer an E.coli Bacterium that expresses streptavidin on its surface and contains a pH sensing promoter that controls the expression of gelatinases. Those bacteria would be able to bind biotinylated nanoparticles. |
| | | | |
| - | <BR><BR>
| + | <BR> |
| - | [[File:Team-EPF-Lausanne Overview-Nanoparticles.jpg|thumb|left|300px|Figure2: Microscope image of gelatine Nanoparticles]] | + | [[File:Team-EPF-Lausanne Overview-Nanoparticles.jpg|thumb|left|300px|Figure 2: Microscope image of gelatine Nanoparticles]] |
| | '''The Nanoparticles'''<BR> | | '''The Nanoparticles'''<BR> |
| | The nanoparticles are made out of gelatin. This kind of nanoparticles has been shown to be able to transport proteins, organic molecules or DNA. They were biotinylated using activated biotin. We invite you to read more about [https://2013.igem.org/Team:EPF_Lausanne/Nanoparticles Nanoparticles]. To assemble the nanoparticles with the bacteria, they only need to be in the same tube, because the interaction between biotin and streptavidin is very strong and spontaneous. | | The nanoparticles are made out of gelatin. This kind of nanoparticles has been shown to be able to transport proteins, organic molecules or DNA. They were biotinylated using activated biotin. We invite you to read more about [https://2013.igem.org/Team:EPF_Lausanne/Nanoparticles Nanoparticles]. To assemble the nanoparticles with the bacteria, they only need to be in the same tube, because the interaction between biotin and streptavidin is very strong and spontaneous. |
| | | | |
| - | <BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR>
| + | <BR><BR><BR><BR><BR><BR><BR><BR> |
| | | | |
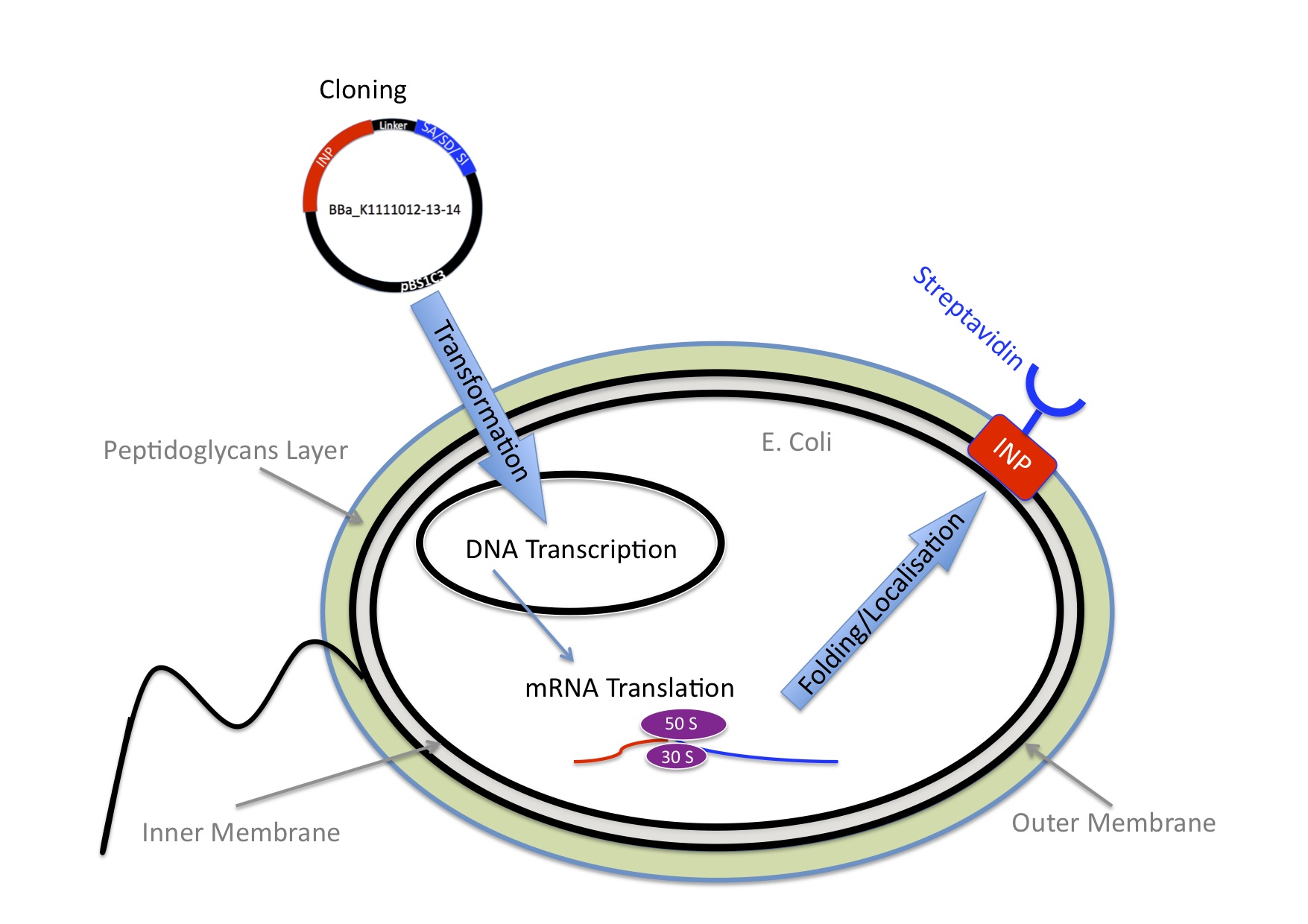
| - | [[Image:EPF-Lausanne-Overview-Coupling.jpg|thumb|left|300px|Figure3: Scheme of Cell surface display]] | + | [[Image:EPF-Lausanne-Overview-Coupling.jpg|thumb|left|300px|Figure 3: Scheme of Cell surface display]] |
| | '''Cell surface display'''<BR> | | '''Cell surface display'''<BR> |
| | To make the bacteria express streptavidin on their surface, we engineered a plasmid that encodes a fusion protein between streptavidin and ice nucleation protein from pseudomonas syringae, which serves as an anchor for the streptavidin to the outer membrane. Read more about [https://2013.igem.org/Team:EPF_Lausanne/Cell_surface_display Cell Surface Display] . At this point the bacteria would be able to transport nanoparticles, but on its own this wouldn’t make any sense. | | To make the bacteria express streptavidin on their surface, we engineered a plasmid that encodes a fusion protein between streptavidin and ice nucleation protein from pseudomonas syringae, which serves as an anchor for the streptavidin to the outer membrane. Read more about [https://2013.igem.org/Team:EPF_Lausanne/Cell_surface_display Cell Surface Display] . At this point the bacteria would be able to transport nanoparticles, but on its own this wouldn’t make any sense. |
| | | | |
| - | <BR><BR><BR><BR><BR><BR><BR><BR><BR>
| + | <BR><BR><BR> |
| | | | |
| - | [[Image:Team-EPF-Lausanne_Overview-Sensing.png|thumb|left|300px|Figure4: Plates of E.coli expressing GFP]] | + | [[Image:Team-EPF-Lausanne_Overview-Sensing.png|thumb|left|300px|Figure 4: Plates of E.coli expressing GFP]] |
| | '''Sensing'''<BR> | | '''Sensing'''<BR> |
| | The crucial point of our project is that E.coli was genetically engineered to sense a specific trigger in the environment. Read more about [https://2013.igem.org/Team:EPF_Lausanne/Sensing-Effector Sensing]. Sensing such a trigger induces the production of enzymes that degrade the nanoparticles, GelE gelatinase from Enterococcus Fecalis and Matrix Metalloprotease MMP2 from H. Sapiens. So the drug will be released exactly where it is needed to treat the patient. | | The crucial point of our project is that E.coli was genetically engineered to sense a specific trigger in the environment. Read more about [https://2013.igem.org/Team:EPF_Lausanne/Sensing-Effector Sensing]. Sensing such a trigger induces the production of enzymes that degrade the nanoparticles, GelE gelatinase from Enterococcus Fecalis and Matrix Metalloprotease MMP2 from H. Sapiens. So the drug will be released exactly where it is needed to treat the patient. |
| | | | |
| - | <BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR>
| |
| | | | |
| - | As mentioned before, we only aimed to make a proof of principle. Eventually, it would be necessary to develop the system in another organism, to add safety mechanisms and develop different sensing plasmid. Read more about the [https://2013.igem.org/Team:EPF_Lausanne/Future Future] applications we imagined. | + | <BR><BR><BR><BR><BR><BR> |
| | + | As mentioned before, we only aimed to make a proof of principle. Eventually, it would be necessary to develop the system in another organism, to add safety mechanisms and develop different sensing plasmids. Read more about the [https://2013.igem.org/Team:EPF_Lausanne/Perspectives future applications] we imagined. |
| | | | |
| | | | |
 "
"