12/09/13
From 2013.igem.org
(Difference between revisions)
TanviSinha (Talk | contribs) |
(→Gel Purification of the pGEM-T pasmid double digestions ran in the 11/09/13) |
||
| (36 intermediate revisions not shown) | |||
| Line 23: | Line 23: | ||
**RFP 1, 2, 3 | **RFP 1, 2, 3 | ||
| + | {|border=1 | ||
| + | |- | ||
| + | |sample||Volume (ul)||Concentration (ng/ul) | ||
| + | |- | ||
| + | |TODF1||43||120 | ||
| + | |- | ||
| + | |TODF2||43||98.8 | ||
| + | |- | ||
| + | |TODX1||42||399.3 | ||
| + | |- | ||
| + | |TODX2||44||442.2 | ||
| + | |- | ||
| + | |TODX3||43||417.5 | ||
| + | |- | ||
| + | |TOBG1||43||262.4 | ||
| + | |- | ||
| + | |TOBG2||47||319.2 | ||
| + | |- | ||
| + | |TOBG3||41||338.4 | ||
| + | |- | ||
| + | |RFP1||41||77.7 | ||
| + | |- | ||
| + | |RFP2||41||32.7 | ||
| + | |- | ||
| + | |RFP3||42||59 | ||
| + | |} | ||
==Glycerol stocks== | ==Glycerol stocks== | ||
| Line 30: | Line 56: | ||
*Samples were then frozen at -80 | *Samples were then frozen at -80 | ||
| + | ==Gel Purification of the pGEM-T pasmid double digestions ran in the [[11/09/13]]== | ||
| + | The gel was purified by using the The Zymoclean Gel DNA recovery Kit and its protocol was followed. | ||
| + | |||
| + | '''Nanodrop Results''' | ||
| + | |||
| + | {|border=1 | ||
| + | |'''Sample'''||'''Volume'''||'''Concentration (ng/ul)'''||'''280/260'''||'''260/230''' | ||
| + | |- | ||
| + | |TODX||48.5||7.1||1.83||0.72 | ||
| + | |- | ||
| + | |TODF||45.5||21.7||1.89||1.19 | ||
| + | |- | ||
| + | |TOBG||47.7||9||1.77||0.70 | ||
| + | |- | ||
| + | |pSB1C3 (RFP)||46.2||7.3||1.81||0.16 | ||
| + | |} | ||
| + | |||
| + | ==Digestion of the samples purified with the enzyme PstI-HF and XbaI== | ||
| + | |||
| + | *Two single digestions were set up to test if one of the enzymes was not working properly. | ||
| + | |||
| + | {|border=1 | ||
| + | |'''DNA volume'''||'''Volume'''||'''Cutsmart buffer'''||'''Enzyme volume'''||'''dH2O added to total volume of 60 ul''' | ||
| + | |- | ||
| + | |TODX||23||6||1||30 | ||
| + | |- | ||
| + | |TODF||22.5||6||1||30.5 | ||
| + | |- | ||
| + | |TOBG||22||6||1||31 | ||
| + | |- | ||
| + | |pSB1C3 (RFP)||20.5||6||1||32.5 | ||
| + | |} | ||
| + | |||
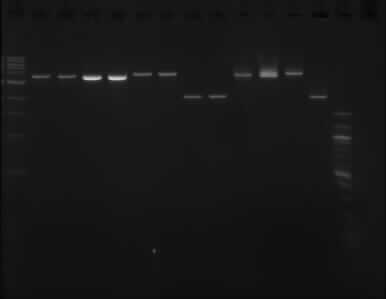
| + | '''Gel image of the above double digestion''' | ||
[[File:doubledigest_gelpurification_12092013.jpg]] | [[File:doubledigest_gelpurification_12092013.jpg]] | ||
| + | |||
| + | Gel order: TODX, TODX, TODF, TODF, TOBG, TOBG, pSB1C3, pSB1C3 (XbaI); TODX, TODF, TOBG, pSB1C3 (PstI) | ||
| + | |||
| + | The picture above shows that the assumption that one of the enzymes did not working properly was wrong, as it shows the same result as the gel run on the [[11/09/13]]. | ||
| + | |||
| + | ==Double digestion of the isolated pGEM plasmids ligated with the TOD operon genes (X, F and TOBG)== | ||
| + | |||
| + | *As the digestions with the plasmids that were stored in the freezer did not work new digestins were set up with the isolated plasmids from the overnight culture. | ||
| + | *The pGEM-T plasmid contain the restrictions sites for PstI and SpeI, which are also present in the amplified TOD operon gene and the pSB1C3 plasmid. | ||
| + | *To avoid the recircularization of the pSB1C3 backbone when ligating the genes with this plasmid, the double digestion was set up with the enzymes PstI and XbaI. | ||
| + | *Another double digestion with EcoRI and SpeI was set up to test if the restrictions sites present in the TOD genes are not mutated. If the bands in the gel shows a linear band, it can be assumed that the enzymes cut the pGEM plasmids in its PstI and SpeI restriction sites. | ||
| + | |||
| + | ===Master Mix of the PstI/XbaI double digestion=== | ||
| + | *30 ul of cut samart buffer | ||
| + | *5 ul of PstI | ||
| + | *5 ul of XbaI | ||
| + | *58 ul of dH2O | ||
| + | **9.8 ul of the master mix were added to each reaction. | ||
| + | **2 ug DNA of each sample were digested. | ||
| + | |||
| + | '''Protocol''' | ||
| + | |||
| + | {|border=1 | ||
| + | |Sample||DNA volume (ul)||dH2O (ul) added to the final volume of 30ul | ||
| + | |- | ||
| + | |TODF1||16.5||3.5 | ||
| + | |- | ||
| + | |TODF2||20.2||0 | ||
| + | |- | ||
| + | |TODX1||5||15.2 | ||
| + | |- | ||
| + | |TODX2||4.2||16 | ||
| + | |- | ||
| + | |TODX3||4.8||15.4 | ||
| + | |- | ||
| + | |TOBG1||7.6||12.6 | ||
| + | |- | ||
| + | |TOBG2||6.3||13.9 | ||
| + | |- | ||
| + | |TOBG3||5.9||14.3 | ||
| + | |} | ||
| + | ===Master Mix of the EcoRI/SpeI double digestion=== | ||
| + | Only samples TODX1, TODX2 and TODX3 were used for this digestion | ||
| + | *15 ul of cutsmart buffer | ||
| + | *2.5 ul of EcoRI | ||
| + | *2.5 ul of SpeI | ||
| + | *106 ul of dH2O | ||
| + | **25 ul of Master Mix was added to each reaction. | ||
| + | |||
| + | {|border=1 | ||
| + | |Sample||DNA volume (ul)||dH2O (ul) added for a final volume of 30 (ul) | ||
| + | |- | ||
| + | |TODX1||5||25 | ||
| + | |- | ||
| + | |TODX2||4.2||0.8 | ||
| + | |- | ||
| + | |TODX3||4.8||0.2 | ||
| + | |} | ||
| + | |||
| + | *The digestion was left overnight in the 37C room. | ||
| + | *Heat kill at 80C for 20 minutes. | ||
| + | |||
| + | ==Chemistry Lab Results== | ||
| + | |||
| + | *Two experiments were run in the chemistry lab. | ||
| + | **Polystyrene Combustion | ||
| + | ***Two pieces of normal packaging polystyrene were burnt, which showed the flammability of polystyrene. | ||
| + | ***After two blocks of polystyrene made form the sugar given to the Leicester iGEM team by Jablite. One of them had DNA incorporated and the other had no DNA. We were able to establish that the polystyrene sugar provided to us from Jablite already had fire retardant chemicals incorporated, which did not allow us to test if DNA could work as a fire retardant for polystyrene. | ||
| + | **Polystyrene dissolution in limonene | ||
Latest revision as of 12:06, 27 September 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Isolate plasmid from overnight culture
- Using Omega mini prep kit protocol
- Samples:
- TODF 1,2,3,
- TODX 1, 2, 3
- TOBG 1, 2, 3,
- RFP 1, 2, 3
| sample | Volume (ul) | Concentration (ng/ul) |
| TODF1 | 43 | 120 |
| TODF2 | 43 | 98.8 |
| TODX1 | 42 | 399.3 |
| TODX2 | 44 | 442.2 |
| TODX3 | 43 | 417.5 |
| TOBG1 | 43 | 262.4 |
| TOBG2 | 47 | 319.2 |
| TOBG3 | 41 | 338.4 |
| RFP1 | 41 | 77.7 |
| RFP2 | 41 | 32.7 |
| RFP3 | 42 | 59 |
Glycerol stocks
- Took 750ul from overnight culture and centrifuged at full speed for ~2 minutes
- Supernatant was removed
- Pellet resuspended in 375ul of HMFM
- Samples were then frozen at -80
Gel Purification of the pGEM-T pasmid double digestions ran in the 11/09/13
The gel was purified by using the The Zymoclean Gel DNA recovery Kit and its protocol was followed.
Nanodrop Results
| Sample | Volume | Concentration (ng/ul) | 280/260 | 260/230 |
| TODX | 48.5 | 7.1 | 1.83 | 0.72 |
| TODF | 45.5 | 21.7 | 1.89 | 1.19 |
| TOBG | 47.7 | 9 | 1.77 | 0.70 |
| pSB1C3 (RFP) | 46.2 | 7.3 | 1.81 | 0.16 |
Digestion of the samples purified with the enzyme PstI-HF and XbaI
- Two single digestions were set up to test if one of the enzymes was not working properly.
| DNA volume | Volume | Cutsmart buffer | Enzyme volume | dH2O added to total volume of 60 ul |
| TODX | 23 | 6 | 1 | 30 |
| TODF | 22.5 | 6 | 1 | 30.5 |
| TOBG | 22 | 6 | 1 | 31 |
| pSB1C3 (RFP) | 20.5 | 6 | 1 | 32.5 |
Gel image of the above double digestion
Gel order: TODX, TODX, TODF, TODF, TOBG, TOBG, pSB1C3, pSB1C3 (XbaI); TODX, TODF, TOBG, pSB1C3 (PstI)
The picture above shows that the assumption that one of the enzymes did not working properly was wrong, as it shows the same result as the gel run on the 11/09/13.
Double digestion of the isolated pGEM plasmids ligated with the TOD operon genes (X, F and TOBG)
- As the digestions with the plasmids that were stored in the freezer did not work new digestins were set up with the isolated plasmids from the overnight culture.
- The pGEM-T plasmid contain the restrictions sites for PstI and SpeI, which are also present in the amplified TOD operon gene and the pSB1C3 plasmid.
- To avoid the recircularization of the pSB1C3 backbone when ligating the genes with this plasmid, the double digestion was set up with the enzymes PstI and XbaI.
- Another double digestion with EcoRI and SpeI was set up to test if the restrictions sites present in the TOD genes are not mutated. If the bands in the gel shows a linear band, it can be assumed that the enzymes cut the pGEM plasmids in its PstI and SpeI restriction sites.
Master Mix of the PstI/XbaI double digestion
- 30 ul of cut samart buffer
- 5 ul of PstI
- 5 ul of XbaI
- 58 ul of dH2O
- 9.8 ul of the master mix were added to each reaction.
- 2 ug DNA of each sample were digested.
Protocol
| Sample | DNA volume (ul) | dH2O (ul) added to the final volume of 30ul |
| TODF1 | 16.5 | 3.5 |
| TODF2 | 20.2 | 0 |
| TODX1 | 5 | 15.2 |
| TODX2 | 4.2 | 16 |
| TODX3 | 4.8 | 15.4 |
| TOBG1 | 7.6 | 12.6 |
| TOBG2 | 6.3 | 13.9 |
| TOBG3 | 5.9 | 14.3 |
Master Mix of the EcoRI/SpeI double digestion
Only samples TODX1, TODX2 and TODX3 were used for this digestion
- 15 ul of cutsmart buffer
- 2.5 ul of EcoRI
- 2.5 ul of SpeI
- 106 ul of dH2O
- 25 ul of Master Mix was added to each reaction.
| Sample | DNA volume (ul) | dH2O (ul) added for a final volume of 30 (ul) |
| TODX1 | 5 | 25 |
| TODX2 | 4.2 | 0.8 |
| TODX3 | 4.8 | 0.2 |
- The digestion was left overnight in the 37C room.
- Heat kill at 80C for 20 minutes.
Chemistry Lab Results
- Two experiments were run in the chemistry lab.
- Polystyrene Combustion
- Two pieces of normal packaging polystyrene were burnt, which showed the flammability of polystyrene.
- After two blocks of polystyrene made form the sugar given to the Leicester iGEM team by Jablite. One of them had DNA incorporated and the other had no DNA. We were able to establish that the polystyrene sugar provided to us from Jablite already had fire retardant chemicals incorporated, which did not allow us to test if DNA could work as a fire retardant for polystyrene.
- Polystyrene dissolution in limonene
- Polystyrene Combustion
 "
"