Template:Kyoto/Notebook/Sep 25
From 2013.igem.org
(Difference between revisions)
(→cDNA Synthesis) |
(→Ligation) |
||
| (14 intermediate revisions not shown) | |||
| Line 172: | Line 172: | ||
<span class="author">nakamoto> | <span class="author">nakamoto> | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||9/24 Pcon-attenuator-aptamer-DT(µL)|| | + | ! ||9/24 Pcon-attenuator-aptamer-DT(µL)||EcoRI(µL)||SpeI(µL)||Buffer(µL)||MilliQ(µL)||total(µL) |
|- | |- | ||
|2 cuts||8.6||1||1||3||16.4||30 | |2 cuts||8.6||1||1||3||16.4||30 | ||
| Line 179: | Line 179: | ||
|} | |} | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! ||9/11 DT(µL)|| | + | ! ||9/11 DT(µL)||EcoRI(µL)||XbaI(µL)||Buffer(µL)||BSA(µL)||MilliQ(µL)||total(µL) |
|- | |- | ||
|2 cuts||10.4||1||1||3||3||11.6||30 | |2 cuts||10.4||1||1||3||3||11.6||30 | ||
| Line 321: | Line 321: | ||
|1||1kbp ladder||--||-- | |1||1kbp ladder||--||-- | ||
|- | |- | ||
| - | |2||Pcon-pT181 attenuator-aptamer 12_1R-DT|| | + | |2||Pcon-pT181 attenuator-aptamer 12_1R-DT||EcoRI||SpeI |
|- | |- | ||
|3||Pcon-pT181 attenuator-aptamer 12_1R-DT||--||-- | |3||Pcon-pT181 attenuator-aptamer 12_1R-DT||--||-- | ||
|- | |- | ||
| - | |4||DT|| | + | |4||DT||EcoRI||XbaI |
|- | |- | ||
|5||DT||--||-- | |5||DT||--||-- | ||
| Line 333: | Line 333: | ||
===Gel Extraction=== | ===Gel Extraction=== | ||
| - | + | ||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">Nakamoto |
{| class="wikitable" | {| class="wikitable" | ||
!Lane||DNA||Enzyme | !Lane||DNA||Enzyme | ||
|- | |- | ||
| - | |1||DT|| | + | |1||DT||EcoRI+XbaI |
|- | |- | ||
| - | |2||DT|| | + | |2||DT||EcoRI+XbaI |
|- | |- | ||
| - | |3||Pcon-pT181 attenuator-aptamer12_1R-DT|| | + | |3||Pcon-pT181 attenuator-aptamer12_1R-DT||EcoRI+SpeI |
|- | |- | ||
| - | |4||Pcon-pT181 attenuator-aptamer12_1R-DT|| | + | |4||Pcon-pT181 attenuator-aptamer12_1R-DT||EcoRI+SpeI |
|- | |- | ||
| - | |5||Pcon-pT181 attenuator-aptamer12_1R-DT|| | + | |5||Pcon-pT181 attenuator-aptamer12_1R-DT||EcoRI+SpeI |
|} | |} | ||
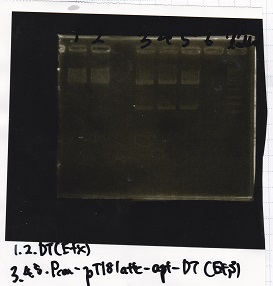
[[File:igku_92514.jpg]]<br> | [[File:igku_92514.jpg]]<br> | ||
| Line 356: | Line 356: | ||
|Pcon-pT181 attenuator-aptamer12_1R-DT(EcoRI&SpeI)||11.8||1.80||0.46 | |Pcon-pT181 attenuator-aptamer12_1R-DT(EcoRI&SpeI)||11.8||1.80||0.46 | ||
|- | |- | ||
| - | |DT( | + | |DT(EcoRI+XbaI)||42.6||1.84||0.98 |
|} | |} | ||
| Line 377: | Line 377: | ||
!state||colspan="2"|Vector||colspan="2"|Inserter||Ligation High ver.2 | !state||colspan="2"|Vector||colspan="2"|Inserter||Ligation High ver.2 | ||
|- | |- | ||
| - | |experiment|| | + | |experiment||9/24 pSB1C3(EcoRI+Spel)||10µL||9/25 PconpT181attenuator-aptamer12-1R-DT(EcoRI+Spel)||5.9µL||8µL |
| - | + | ||
| - | + | ||
|} | |} | ||
</div> | </div> | ||
| Line 385: | Line 383: | ||
===PCR=== | ===PCR=== | ||
| - | |||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">Hirano |
{| class="wikitable" | {| class="wikitable" | ||
!800pg/μL DT||KOD plus||10x buffer||dNTP||MgSO4||1C3_GG2_fwd||iC3_GG1_rev||MilliQ||total | !800pg/μL DT||KOD plus||10x buffer||dNTP||MgSO4||1C3_GG2_fwd||iC3_GG1_rev||MilliQ||total | ||
| Line 402: | Line 399: | ||
===Transformation=== | ===Transformation=== | ||
| - | |||
<div class="experiment"> | <div class="experiment"> | ||
<span class="author">Tatsui</span> | <span class="author">Tatsui</span> | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | !Name | + | !Name||Plate |
|- | |- | ||
| - | |aptamer12-1R-DT | + | |aptamer12-1R-DT||CP |
|- | |- | ||
| - | |pSB1C3(EcoRI+SpeI)+pT181attenuator(EcoRI+Spal) | + | |pSB1C3(EcoRI+SpeI)+pT181attenuator(EcoRI+Spal)||CP |
|- | |- | ||
| - | |Pcon-pT181attenuator(EcoRI+XbaI)+RBS-GFP-DT(EcoRI+SpeI) | + | |Pcon-pT181attenuator(EcoRI+XbaI)+RBS-GFP-DT(EcoRI+SpeI)||Amp |
|- | |- | ||
| - | |Pcon-RBS-GFP-DT(EcoRI+XbaI)+Pcon-RBS-tetR-DT(EcoRI+SpeI) | + | |Pcon-RBS-GFP-DT(EcoRI+XbaI)+Pcon-RBS-tetR-DT(EcoRI+SpeI)||Amp |
|- | |- | ||
| - | |pSB1C3(EcoRI+SpeI)+Pcon-pT181attenuator-aptamer12-1R-DT(EcoRI+SpeI) | + | |pSB1C3(EcoRI+SpeI)+Pcon-pT181attenuator-aptamer12-1R-DT(EcoRI+SpeI)||CP |
|} | |} | ||
</div> | </div> | ||
| - | |||
===Electrophoresis=== | ===Electrophoresis=== | ||
| Line 449: | Line 444: | ||
Concentrate PCR product by evaporator until volume become less than 3µL, add all sulution and pipetting. </div> | Concentrate PCR product by evaporator until volume become less than 3µL, add all sulution and pipetting. </div> | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | ! | + | !9/25 DT(2-1)||10*cut smart Buffer||BsaI-HF||MilliQ||total |
|- | |- | ||
| - | + | |3.0 µL||2.0 µL||0.3 µL||14.7 µL||20.0 µL | |
|} | |} | ||
Incubate 37°C 12hour | Incubate 37°C 12hour | ||
| - | |||
===Liquid Culture=== | ===Liquid Culture=== | ||
| - | |||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">Hirano</span> |
{| class="wikitable" | {| class="wikitable" | ||
!Sample||medium | !Sample||medium | ||
| Line 570: | Line 563: | ||
|RNase-frec water||4 µL | |RNase-frec water||4 µL | ||
|} | |} | ||
| - | |||
Incubate 42°C 2min, then on ice immediately<BR> | Incubate 42°C 2min, then on ice immediately<BR> | ||
Add <BR> | Add <BR> | ||
| Line 580: | Line 572: | ||
|RT Primer Mix||0.5µL | |RT Primer Mix||0.5µL | ||
|} | |} | ||
| - | |||
Incubate 42°C 15min, then incubate 95°C 3min | Incubate 42°C 15min, then incubate 95°C 3min | ||
| + | </div> | ||
===Electrophoresis=== | ===Electrophoresis=== | ||
| Line 600: | Line 592: | ||
===Ligation=== | ===Ligation=== | ||
| - | + | ||
| - | + | ||
<div class="experiment"> | <div class="experiment"> | ||
| - | <span class="author"> | + | <span class="author">Tatsui</span> |
{| class="wikitable" | {| class="wikitable" | ||
!state||colspan="2"|Vector||colspan="2"|Inserter||Ligation High ver.2 | !state||colspan="2"|Vector||colspan="2"|Inserter||Ligation High ver.2 | ||
| Line 610: | Line 601: | ||
|} | |} | ||
</div> | </div> | ||
| - | |||
Latest revision as of 19:26, 27 September 2013
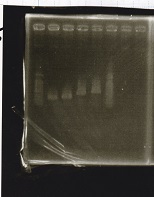
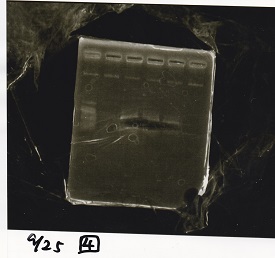
Sep 25
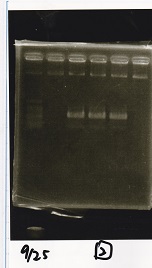
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | 100bp ladder | -- | -- |
| 2 | Pcon-antisense (1C3-66E-fwd A2-662 rev) | -- | -- |
| 3 | DT (1C3-661-fwd 1C3 662 rev) | -- | -- |
| 4 | spinach-DT(1C3-661-fwd 1C3 662 rev) | -- | -- |
| 5 | Pcon-antisense-spinach-DT(1C3-66E fwd A2-661 rev) | -- | -- |
| 6 | 100bp ladder | -- | -- |
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 2 | 100bp ladder | -- | -- |
| 3 | Ptet (1C3 66E fwd-1C3 660 rev) | -- | -- |
| 4 | Pcon-attenuator(1C3-660 fwd A2-661 rev) | -- | -- |
| 5 | DT(1C3 662 fwd-1C3 661 rev) | -- | -- |
| 6 | Pcon-attenuator-aptamer-DT(1C3 66E fwd-A2-661-rev) | -- | -- |
| 7 | 100bp ladder | -- | -- |
Colony PCR
| Sample | base pair |
|---|---|
| 9/23 Pcon-PT181 attenuator-aptamer12-1R-DT-(5~12) | 859 |
| 9/23 Pcon-PT181 attenuator-RBS-GFP-DT-(5~8) | 1527 |
| 9/23 Pcon-PT181 attenuator-pSB1C3-(5~8) | 601 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94 °C | 94 °C | 55 °C | 68 °C | -- |
| 2 min | 30 s | 30 s | 1min5s | 30 cycles |
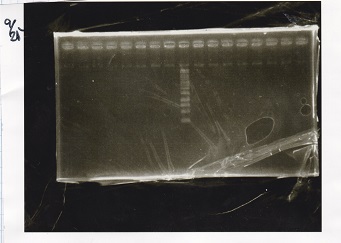
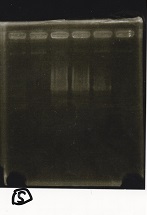
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | Pcon-PT181 attenuator-aptamer12-1R-DT-5 | -- | -- |
| 2 | Pcon-PT181 attenuator-aptamer12-1R-DT-6 | -- | -- |
| 3 | Pcon-PT181 attenuator-aptamer12-1R-DT-7 | -- | -- |
| 4 | Pcon-PT181 attenuator-aptamer12-1R-DT-8 | -- | -- |
| 5 | Pcon-PT181 attenuator-aptamer12-1R-DT-9 | -- | -- |
| 6 | Pcon-PT181 attenuator-aptamer12-1R-DT-10 | -- | -- |
| 7 | Pcon-PT181 attenuator-aptamer12-1R-DT-11 | -- | -- |
| 8 | Pcon-PT181 attenuator-aptamer12-1R-DT-12 | -- | -- |
| 9 | 1kbp ladder | -- | -- |
| 10 | Pcon-PT181 attenuator-RBS-GFP-DT-5 | -- | -- |
| 11 | Pcon-PT181 attenuator-RBS-GFP-DT-6 | -- | -- |
| 12 | Pcon-PT181 attenuator-RBS-GFP-DT-7 | -- | -- |
| 13 | Pcon-PT181 attenuator-RBS-GFP-DT-8 | -- | -- |
| 14 | Pcon-PT181 attenuator-pSB1C3-5 | -- | -- |
| 15 | Pcon-PT181 attenuator-pSB1C3-6 | -- | -- |
| 16 | Pcon-PT181 attenuator-pSB1C3-7 | -- | -- |
| 17 | Pcon-PT181 attenuator-pSB1C3-8 | -- | -- |
Transformation
| Name | Sample | Competent Cells | Total | Plate |
|---|---|---|---|---|
| Plac+tet aptamer12+Pcon tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-pT181attenuator-DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-tet aptamer12-DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-Spinach-DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-pT181antisense+DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-pT181attenuator+tet aptamer12-DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon tetR-DT+Pcon-GFP-DT+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-tet aptamer12-DT+Pcon-pT181attenuator+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-pT181attenuator-DT+Pcon-pT181attenuator+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-tet aptamer12-DT+Ptet-pT181antisense+Spinach-DT+Pcon-RBS-GFP-DT+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-tet aptamer12-DT+Pcon-pT181antisense+Spinach-DT+Pcon-pT181attenuator+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Pcon-tet aptamer12-DT+Ptet-pT181antisense+Spinach-DT+Pcon-pT181attenuator+RBS-GFP-DT | 1µL | 10µL | 11µL | -- |
| Name | Sample | Competent Cells | Total | Plate |
|---|---|---|---|---|
| aptamer12-DT(EcoRI+XbaI)+Pcon-pT181attenuator(EcoRI+SpeI) | 2µL | 20µL | 22µL | -- |
| Ptet(SpeI+PstI)+RBS-GFP-DT(XbaI+PstI)) | 2µL | 20µL | 22µL | -- |
Ligation
| state | Vector | Inserter | Ligation High ver.2 | ||
|---|---|---|---|---|---|
| experiment | 9/25 aptamer12_1R-DT(EcoRI&XbaI) | 4.5 µL | Pcon-pT181 attenuator(EcoRI&SpeI) | 7.7 µL | 6.1 µL |
| experiment | 9/17 pSB1C3(EcoRI&SpeI) | 2.0 µL | 9/3 pT181 attenuator(EcoRI&SpeI) | 14 µL | 8 µL |
| experiment | 9/25 Pcon-pT181 attenuator(EcoRI&XbaI) | 4.5 µL | 9/25 RBS-GFP-DT(EcoRI&SpeI) | 19.3 µL | 11.9 µL |
| experiment | 9/25 Pcon-RBS-GFP-DT(EcoRI&XbaI) | 4.2 µL | Pcon-RBS-tetR-DT(EcoRI&SpeI) | 20 µL | 12.1 µL |
Restriction Enzyme Digestion
| 9/24 Pcon-attenuator-aptamer-DT(µL) | EcoRI(µL) | SpeI(µL) | Buffer(µL) | MilliQ(µL) | total(µL) | |
|---|---|---|---|---|---|---|
| 2 cuts | 8.6 | 1 | 1 | 3 | 16.4 | 30 |
| NC | 0.4 | 0 | 0 | 1 | 8.6 | 10 |
| 9/11 DT(µL) | EcoRI(µL) | XbaI(µL) | Buffer(µL) | BSA(µL) | MilliQ(µL) | total(µL) | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 10.4 | 1 | 1 | 3 | 3 | 11.6 | 30 |
| NC | 0.5 | 0 | 0 | 1 | 1 | 7.5 | 10 |
Liquid Culture
| Sample | medium |
|---|---|
| DT | -- |
| pT181attenuator | -- |
| Pcon-RBS-tetR-DT | -- |
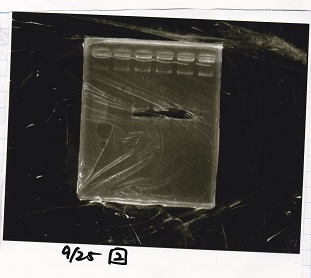
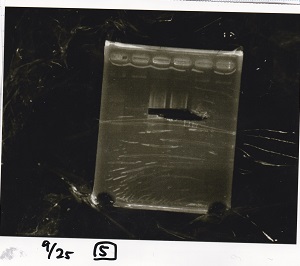
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | -- |
| 2 | Pcon-pT181antisense | -- |
| 3 | ||
| 4 | ||
| 5 | -- | -- |
| 6 | DT | -- |
| 7 | ||
| 8 | ||
| 9 | -- | -- |
| 10 | Spinach-DT | -- |
| 11 | ||
| 12 |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | -- |
| 3 | Pcon-antisense-spinach-DT(E-1A) | -- |
| 4 | Pcon-antisense-spinach-DT(E-1A) | -- |
| 5 | Pcon-antisense-spinach-DT(E-1A) | -- |
| Lane | DNA | Enzyme |
|---|---|---|
| 7 | 100bp ladder | -- |
| 9 | Ptet(E-0) | -- |
| 10 | Ptet(E-0) | -- |
| 11 | Ptet(E-0) | -- |
| Lane | DNA | Enzyme |
|---|---|---|
| 13 | 100bp ladder | -- |
| 15 | Pcon-attenuator(0-1A) | -- |
| 16 | Pcon-attenuator(0-1A) | -- |
| 17 | Pcon-attenuator(0-1A) | -- |
| Lane | DNA | Enzyme |
|---|---|---|
| 19 | 100bp ladder | -- |
| 21 | Pcon-attenuator-aptamer-DT(E-1A) | -- |
| 22 | Pcon-attenuator-aptamer-DT(E-1A) | -- |
| 23 | Pcon-attenuator-aptamer-DT(E-1A) | -- |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| DT(2-1) | 24.9 | 1.73 | 0.37 |
| Ptet(E-0) | 12.8 | 1.59 | 0.90 |
| Pcon-antisense(E-2A) | 16.5 | 1.72 | 1.07 |
| Pcon-attenuator-aptamer-DT(E-1A) | 22.1 | 1.75 | 0.94 |
| Pcon-antisense-spinach-DT(E-1A) | 31.0 | 1.78 | 1.15 |
| spinach-DT(1-2) | 23.4 | 1.85 | 1.11 |
| Pcon-attenuator(0-1A) | 22.5 | 1.76 | 0.46 |
| DT(1-2) | 25.1 | 1.78 | 1.21 |
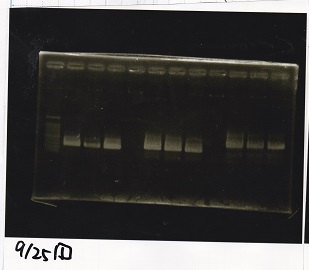
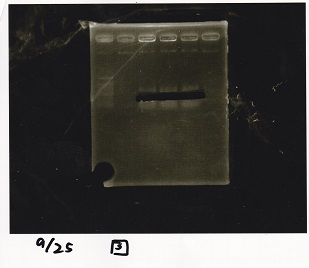
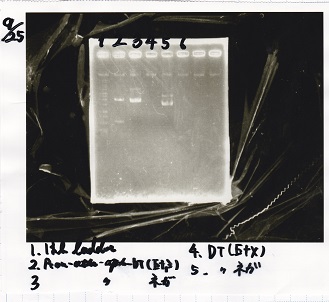
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | 1kbp ladder | -- | -- |
| 2 | Pcon-pT181 attenuator-aptamer 12_1R-DT | EcoRI | SpeI |
| 3 | Pcon-pT181 attenuator-aptamer 12_1R-DT | -- | -- |
| 4 | DT | EcoRI | XbaI |
| 5 | DT | -- | -- |
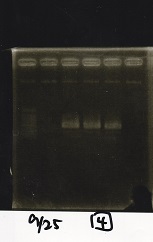
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | DT | EcoRI+XbaI |
| 2 | DT | EcoRI+XbaI |
| 3 | Pcon-pT181 attenuator-aptamer12_1R-DT | EcoRI+SpeI |
| 4 | Pcon-pT181 attenuator-aptamer12_1R-DT | EcoRI+SpeI |
| 5 | Pcon-pT181 attenuator-aptamer12_1R-DT | EcoRI+SpeI |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-pT181 attenuator-aptamer12_1R-DT(EcoRI&SpeI) | 11.8 | 1.80 | 0.46 |
| DT(EcoRI+XbaI) | 42.6 | 1.84 | 0.98 |
Liquid Culture
| Sample | medium |
|---|---|
| Pcon-pT181anntisense-spinach-DT(4k5) | - |
Ligation
| state | Vector | Inserter | Ligation High ver.2 | ||
|---|---|---|---|---|---|
| experiment | 9/24 pSB1C3(EcoRI+Spel) | 10µL | 9/25 PconpT181attenuator-aptamer12-1R-DT(EcoRI+Spel) | 5.9µL | 8µL |
PCR
| 800pg/μL DT | KOD plus | 10x buffer | dNTP | MgSO4 | 1C3_GG2_fwd | iC3_GG1_rev | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 57°C | 68°C | -- |
| 2min | 10s | 30s | 24s | 30cycles |
Transformation
| Name | Plate |
|---|---|
| aptamer12-1R-DT | CP |
| pSB1C3(EcoRI+SpeI)+pT181attenuator(EcoRI+Spal) | CP |
| Pcon-pT181attenuator(EcoRI+XbaI)+RBS-GFP-DT(EcoRI+SpeI) | Amp |
| Pcon-RBS-GFP-DT(EcoRI+XbaI)+Pcon-RBS-tetR-DT(EcoRI+SpeI) | Amp |
| pSB1C3(EcoRI+SpeI)+Pcon-pT181attenuator-aptamer12-1R-DT(EcoRI+SpeI) | CP |
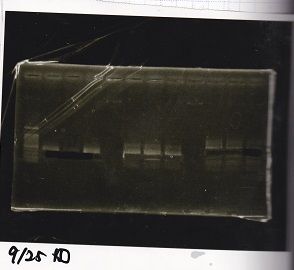
Electrophoresis
Column Refining
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| 8/25 DT(2-1) | -- | -- | -- |
Restriction Enzyme Digestion
Concentrate PCR product by evaporator until volume become less than 3µL, add all sulution and pipetting.
| 9/25 DT(2-1) | 10*cut smart Buffer | BsaI-HF | MilliQ | total |
|---|---|---|---|---|
| 3.0 µL | 2.0 µL | 0.3 µL | 14.7 µL | 20.0 µL |
Incubate 37°C 12hour
Liquid Culture
| Sample | medium |
|---|---|
| Pcon-GFP-DT | LB(Amp) |
| RBS-GFP-DT | LB(CP) |
| Pcon-spinach-DT | LB(Amp) |
| Pcon-tetRaptamer-DT | LB(Amp) |
| Pcon-attenuator-DT | LB(Amp) |
| spinach-DT | LB(CP) |
| Pcon-antisense-spinach-DT | LB(Amp) |
Plating
Mix Liquid Culture 1mL and LB(+Amp), incubate 37°C 30min
| Sample | Use plate | The number of plate |
|---|---|---|
| Pcon-GFP-DT | M9(+Amp) | 3 |
| Pcon-GFP-DT | LB(+Amp) | 3 |
</div> incubate 37°C
Colony PCR
| Sample | base pair |
|---|---|
| 9/24 GGA-1(1~8) | 3177 |
| 9/24 GGA-2(1~8) | 3189 |
| 9/24 GGA-3(1~8) | 2989 |
| 9/24 GGA-4(1~8) | 2939 |
| 9/24 GGA-7(1) | 2489 |
| GGA-16(1) | 2717 |
| RBS-GFP-DT | -- |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 3min15s | 30cycles |
RNA Extraction
sumple 0.25mL
Add 1mLISOGEN-LS
Lysis or homogenization
Store for 5 min, at room temperature
Add 0.2mL Chloroform
Shake vigorously for 15sec.
Store for 2~3min.at room temperature
Centrifuge 12K*g for 15min. at 4°C
Extract aqueous phase
Add 0.5mL isopropanol
Store for 5~10min. at room temperature
Centrifuge 12K*g for 10min. at 4°C
Remove aqueous phase
Add at least 1mL 70% ethanol
Vortex
Centrifuge 7.5K*g for 5min. at 4°C
Remove aqueous phase
Dry briefly
Add ddH2O, TE(pH8.0) or 0.5% SDS
Dissolve
Total RNA solution
| DNA | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-RBS-GFP-DT | 2783 | 1.95 | 1.48 |
Add 91.2µL, Store at freezer
Total 250µL</div>
cDNA Synthesis
| volume | 7 µL |
|---|---|
| 7*cDNA Wipcout Buffer | 1 µL |
| Template RNA(9/25 Pcon-RBS-GFP-DT) | 3 µL |
| RNase-frec water | 4 µL |
Incubate 42°C 2min, then on ice immediately
Add
| 5*Quantiscript Reverse Transcriptase | 0.5 µL |
|---|---|
| 5*Quantiscript RT Buffer | 2 µL |
| RT Primer Mix | 0.5µL |
Incubate 42°C 15min, then incubate 95°C 3min
Electrophoresis
Ligation
| state | Vector | Inserter | Ligation High ver.2 | ||
|---|---|---|---|---|---|
| experiment | 9/13 Pcon-pT181-attenuator(SpeI&PstI) | 3.5µL | 9/25 RBS-GFP-DT(XbaI&PstI) | 13.5µL | 8.5 µL |
 "
"