Team:UCLA/Project/NaturalSystem
From 2013.igem.org
Michaelc1618 (Talk | contribs) |
|||
| (9 intermediate revisions not shown) | |||
| Line 38: | Line 38: | ||
<li><a href="/Team:UCLA/Modeling">MODELING</a></li> | <li><a href="/Team:UCLA/Modeling">MODELING</a></li> | ||
<li><a href="/Team:UCLA/HumanPractices">HUMAN PRACTICES</a></li> | <li><a href="/Team:UCLA/HumanPractices">HUMAN PRACTICES</a></li> | ||
| - | <li><a href="/Team:UCLA/Notebook">NOTEBOOK</a></li> | + | <li><a href="/Team:UCLA/Notebook/Biobrick">NOTEBOOK</a></li> |
<li><a href="/Team:UCLA/Safety">SAFETY</a></li> | <li><a href="/Team:UCLA/Safety">SAFETY</a></li> | ||
<li><a href="/Team:UCLA/Attributions">ATTRIBUTIONS</a></li> | <li><a href="/Team:UCLA/Attributions">ATTRIBUTIONS</a></li> | ||
| Line 74: | Line 74: | ||
</div> | </div> | ||
| + | <div> | ||
<h4>Diversity Generation</h4> | <h4>Diversity Generation</h4> | ||
| + | [[File:Natural_DGR_systems.PNG|thumb|left|The genomic DGR system in BPP-1 and other phages (Medhekar)|500px]] | ||
<p>The BPP-1 phage contains a diversity generating retroelement (DGR) for diversifying its Mtd protein. DGRs generate variability in the viral receptor protein that specifies tropism for ligand molecules, and encode polypeptides that are designed to tolerate sequence variation. A C-type lectin fold displays amino acid sequence variability. The lectin fold provides a static scaffold that helps balance diversity with protein stability. A reverse transcriptase mediates the exchange between two repeats, a variable repeat (VR) and a template repeat (TR). </p> | <p>The BPP-1 phage contains a diversity generating retroelement (DGR) for diversifying its Mtd protein. DGRs generate variability in the viral receptor protein that specifies tropism for ligand molecules, and encode polypeptides that are designed to tolerate sequence variation. A C-type lectin fold displays amino acid sequence variability. The lectin fold provides a static scaffold that helps balance diversity with protein stability. A reverse transcriptase mediates the exchange between two repeats, a variable repeat (VR) and a template repeat (TR). </p> | ||
<p>The BPP-1 genome contains a variable repeat (VR) of 134 base pairs on the gene coding for the Mtd. This variable repeat is variable across every specimen of BPP-1, with variations isolated to 23 discrete nucleotides. Downstream from the variable repeat is the template repeat (TR), which serves as a master copy and is never permanently altered in wild-type phage (Medhekar). </p> | <p>The BPP-1 genome contains a variable repeat (VR) of 134 base pairs on the gene coding for the Mtd. This variable repeat is variable across every specimen of BPP-1, with variations isolated to 23 discrete nucleotides. Downstream from the variable repeat is the template repeat (TR), which serves as a master copy and is never permanently altered in wild-type phage (Medhekar). </p> | ||
<p>Occasionally, during the viral assembly process, the TR is transcribed, and then reverse transcribed by a reverse transcriptase coded for by the <i>brt</i> gene further downstream from the <i>mtd</i> gene. This process introduces site mutations at the 23 extremely specific nucleotide locations. These nucleotides are all adenines, and are changed to any of the four possible nucleotides. The resulting cDNA strand is then directed to the location of the VR and integrated into the genome, replacing the VR itself (Guo). All of this results in the assembled viral particle having a different sequence in the <i>mtd</i> gene, and thus a different Mtd protein that is capable of binding to another target on the surface of <i>Bordetella</i>.</p> | <p>Occasionally, during the viral assembly process, the TR is transcribed, and then reverse transcribed by a reverse transcriptase coded for by the <i>brt</i> gene further downstream from the <i>mtd</i> gene. This process introduces site mutations at the 23 extremely specific nucleotide locations. These nucleotides are all adenines, and are changed to any of the four possible nucleotides. The resulting cDNA strand is then directed to the location of the VR and integrated into the genome, replacing the VR itself (Guo). All of this results in the assembled viral particle having a different sequence in the <i>mtd</i> gene, and thus a different Mtd protein that is capable of binding to another target on the surface of <i>Bordetella</i>.</p> | ||
| + | </div> | ||
<h4>Comparable Systems in Nature</h4> | <h4>Comparable Systems in Nature</h4> | ||
| - | <p> | + | <p>The human immune system can generate upwards of 10<sup>12</sup> antibodies with unique antigen-binding sites. It is remarkable that such enormous diversity can be generated from far fewer numbers of genes, and does not disrupt the overall structure of the antibody. This strategy of shuffling and combining parts has been tried-and-true throughout the tree of life, and has conferred an adaptive advantage to many species – including ourselves! Evolution trends towards making the most out of a handful of tools, and it’s no wonder that we are interested in upholding this principle for our own projects (Alberts).</p> |
| - | < | + | |
| + | <html> | ||
<div id="reference"> | <div id="reference"> | ||
<b>REFERENCES</b> | <b>REFERENCES</b> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | <p> | + | <p>Alberts, B., et al. <a href="http://www.ncbi.nlm.nih.gov/books/NBK26860/">"The Generation of Antibody Diversity."</a> ''Molecular Biology of the Cell.'' New York: Garland Science, 2002. Print.</p> |
| - | <p> | + | <p>Dai, W., et al. <a href="http://www.ncbi.nlm.nih.gov/pubmed/20160083">"Three-dimensional structure of tropism-switching <i>Bordetella</i> bacteriophage."</a> <i>PNAS</i>. 107.9 (2010):4347-52. Print.</p> |
| + | |||
| + | <p>Guo, H., et al. <a href="http://www.ncbi.nlm.nih.gov/pubmed/22194701">"Target Site Recognition by a Diversity-Generating Retroelement."</a> <i>PLoS Genetics</i>. 7.12 (2011). Web.</p> | ||
| + | |||
| + | <p>Liu, M., et al. <a href="http://www.sciencemag.org/content/295/5562/2091">"Reverse Transcriptase-Mediated Tropism Switching in Bordetella Bacteriophage."</a> <i>Science</i>. 295.5562 (2002): 2091-4. Print.</p> | ||
| + | |||
| + | <p>Medhekar, B., J. Miller. <a href="http://www.ncbi.nlm.nih.gov/pubmed/17703991">"Diversity-Generating Retroelements."</a> <i>Curr Opin Microbiol</i>. 10.4 (2007):388-95. Print.</p> | ||
| + | |||
| + | <p><a href="http://viralzone.expasy.org/all_by_species/141.html">"Podoviridae."</a> <i>ViralZone</i>. ExPASy, n.d. Web. 2013.</p> | ||
| + | </html> | ||
Latest revision as of 02:13, 28 September 2013
Contents |
The Natural Host
Bordetella is the natural host of the BPP-1 bacteriophage. It expresses a protein on its surface, pertactin, that BPP-1 binds to in the first step of the infection process. However, Bordetella does not always express pertactin. It cycles between two phases: Bvg+ and Bvg-. Pertactin is only expressed in the Bvg+ phase, while in the Bvg- phase, pertactin expression is inhibited. Infection by BPP-1 during this phase is much less common, but it still occurs (Liu). This indicates that the BPP-1 virus has a mechanism for changing its host specificity, and thus, the binding properties of the proteins on its tail fibers.
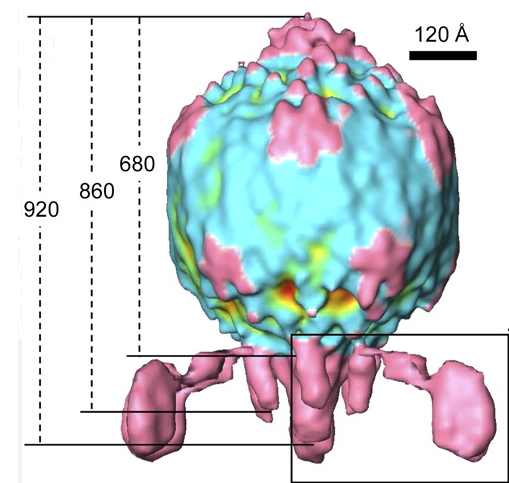
The Virus
The BPP-1 virus is a bacteriophage belonging to the Podoviridae family. It has an icosahedral head with T=7 symmetry, and a short, noncontractile tail with six tail “spikes” attached to tail fibers. At the end of these tail fibers are major tropism determinant (Mtd) proteins, which bind to the pertactin proteins expressed on the surface of Bordetella. The Mtd protein does not display high affinity for pertactin, but the multiple Mtd proteins possessed by each phage particle coupled with the flexibility of the tail fibers allow BPP-1 as a whole to have high avidity for its host (Liu). Following infection, the virus is lytic, and destroys the bacterial host to release more copies of itself (ViralZone).
Diversity Generation
The BPP-1 phage contains a diversity generating retroelement (DGR) for diversifying its Mtd protein. DGRs generate variability in the viral receptor protein that specifies tropism for ligand molecules, and encode polypeptides that are designed to tolerate sequence variation. A C-type lectin fold displays amino acid sequence variability. The lectin fold provides a static scaffold that helps balance diversity with protein stability. A reverse transcriptase mediates the exchange between two repeats, a variable repeat (VR) and a template repeat (TR).
The BPP-1 genome contains a variable repeat (VR) of 134 base pairs on the gene coding for the Mtd. This variable repeat is variable across every specimen of BPP-1, with variations isolated to 23 discrete nucleotides. Downstream from the variable repeat is the template repeat (TR), which serves as a master copy and is never permanently altered in wild-type phage (Medhekar).
Occasionally, during the viral assembly process, the TR is transcribed, and then reverse transcribed by a reverse transcriptase coded for by the brt gene further downstream from the mtd gene. This process introduces site mutations at the 23 extremely specific nucleotide locations. These nucleotides are all adenines, and are changed to any of the four possible nucleotides. The resulting cDNA strand is then directed to the location of the VR and integrated into the genome, replacing the VR itself (Guo). All of this results in the assembled viral particle having a different sequence in the mtd gene, and thus a different Mtd protein that is capable of binding to another target on the surface of Bordetella.
Comparable Systems in Nature
The human immune system can generate upwards of 1012 antibodies with unique antigen-binding sites. It is remarkable that such enormous diversity can be generated from far fewer numbers of genes, and does not disrupt the overall structure of the antibody. This strategy of shuffling and combining parts has been tried-and-true throughout the tree of life, and has conferred an adaptive advantage to many species – including ourselves! Evolution trends towards making the most out of a handful of tools, and it’s no wonder that we are interested in upholding this principle for our own projects (Alberts).
Alberts, B., et al. "The Generation of Antibody Diversity." ''Molecular Biology of the Cell.'' New York: Garland Science, 2002. Print.
Dai, W., et al. "Three-dimensional structure of tropism-switching Bordetella bacteriophage." PNAS. 107.9 (2010):4347-52. Print.
Guo, H., et al. "Target Site Recognition by a Diversity-Generating Retroelement." PLoS Genetics. 7.12 (2011). Web.
Liu, M., et al. "Reverse Transcriptase-Mediated Tropism Switching in Bordetella Bacteriophage." Science. 295.5562 (2002): 2091-4. Print.
Medhekar, B., J. Miller. "Diversity-Generating Retroelements." Curr Opin Microbiol. 10.4 (2007):388-95. Print.
"Podoviridae." ViralZone. ExPASy, n.d. Web. 2013.
 "
"