Team:Tsinghua/Project-Sensor
From 2013.igem.org
| (6 intermediate revisions not shown) | |||
| Line 108: | Line 108: | ||
</div> | </div> | ||
<script type="text/javascript"> | <script type="text/javascript"> | ||
| - | + | initialize(); | |
| - | + | setupMenu(2, 1); | |
| - | + | </script> | |
</div> | </div> | ||
</div> | </div> | ||
<div id="mycontent"> | <div id="mycontent"> | ||
<div class="normal"> | <div class="normal"> | ||
| - | <h1>PPD Sensor</h1> | + | <div class="normal"> |
| - | <h2>Overview</h2> | + | <h1>PPD Sensor</h1> |
| - | <p> | + | <div id="brief"> |
| - | + | <p> | |
| - | + | We created <b>PPD sensor</b> part based on <b>quorum sensing system</b> in yeast. The sensor yeast can sense the <b>AHL</b> secreted by bacteria in surroundings. After AHL inducing, the reporter gene mCherry in sensor system can be expressed successfully. | |
| - | <p> | + | </p> |
| - | + | </div> | |
| - | + | ||
| - | <p> | + | <div id="section-nav"> |
| - | + | <ul> | |
| - | + | <li class="section1"> | |
| - | + | Overview | |
| - | + | </li> | |
| - | + | <li class="section2"> | |
| - | + | Design | |
| + | </li> | ||
| + | <li class="section3"> | ||
| + | Experiments and Results | ||
| + | </li> | ||
| + | <li class="section4"> | ||
| + | Discussion | ||
| + | </li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | |||
| + | <div class="section section1"> | ||
| + | <h2>Overview</h2> | ||
| + | <p> | ||
| + | We constructed the PPD sensor part by reconstructing bacteria quorum sensing system in yeast. | ||
| + | </p> | ||
| + | <p> | ||
| + | We confirm that the Lux Receptor in the sensor was expressed. | ||
| + | </p> | ||
| + | <p> | ||
| + | We realized the inducing regulation of our sensor part by adding AHL to the yeast. | ||
| + | </p> | ||
<div class="figure"> | <div class="figure"> | ||
| - | <img class="center" src="https://static.igem.org/mediawiki/2013/ | + | <img class="center" src="https://static.igem.org/mediawiki/2013/e/eb/Tsinghua-sensor-1.png"/> |
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 1. Overview of PPD sensor | |
| - | + | </p> | |
</div> | </div> | ||
| - | <h2> | + | </div> |
| - | <p> | + | |
| - | + | ||
| - | + | ||
| - | <div class="figure"> | + | <div class="section section2"> |
| - | <img class="center" src="https://static.igem.org/mediawiki/2013/ | + | <h2>Design</h2> |
| + | |||
| + | <h3>Designed PPD sensor part</h3> | ||
| + | <p> | ||
| + | To achieve the sensing of the AHLs from the pathogenic organisms, we plan to introduce the <b>quorum sensing</b> system to yeast, which has been naturally used by the <b>pathogenic microoganisms</b> themselves for communicating within the colonies. Usually, the quorum sensing system share the similar working mechanisms between different species, the precise procedures are shown as follows: | ||
| + | </p> | ||
| + | <div class="figure"> | ||
| + | <img class="center" src="https://static.igem.org/mediawiki/2013/d/dd/Tsinghua-sensor-002.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 2. Principle of quorum sensing system | |
| - | + | </p> | |
</div> | </div> | ||
| - | < | + | <h3>Reconstruction the PPD system</h3> |
| - | + | <p> | |
| - | + | Considered the difference between <b>prokaryotic</b> and <b>eukaryotic</b> system, the prokaryotic quorum sensing system needs to be reconstructed in a “eukaryotic way” when introduced into yeast. As the quorum sensing systems in different species share the similar mechanism, we typically reconstructed the <b>LuxR</b> system. | |
| - | <p> | + | </p> |
| - | + | <div class="figure"> | |
| - | + | <img class="center" src="https://static.igem.org/mediawiki/2013/0/0d/Tsinghua-sensor-3.png"/> | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | <div class="figure"> | + | |
| - | <img class="center" src="https://static.igem.org/mediawiki/2013/0/ | + | |
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 3. Reconstruction of quorum sensing system in yeast | |
| - | + | </p> | |
</div> | </div> | ||
| - | < | + | <p> |
| - | < | + | For the reconstruction of LuxR, we add three times repeat of <b>VP16</b> region, as well as a region of <b>NLS</b>. The 3 times repeated sequence of VP16 is function as activation domain (AD) to link the RNA POL II, the Nuclear Localization Sequence (NLS) is aimed to import the LuxR to the nuclear and thus enhance the transcriptional activating potency. |
| - | + | </p> | |
| - | + | <p> | |
| - | <div class="figure"> | + | For the reconstruction of <b>Lux</b> Promoter, we add <b>cyc100 mini promoter (the TATA Box)</b>, the function of which is to recruit RNA POL II to DNA region. |
| - | <img class="center" src="https://static.igem.org/mediawiki/2013/ | + | </p> |
| + | <p> | ||
| + | These modifications will lead to the effect that when AHL is introduced into the cell, LuxR will bind with AHL and then together bind with Lux promoter, the VP16 domain of the complex will then bind with POL II, which bind to <b>cyc100 mini promoter</b> and facilitate the gene expression downstream of <b>plux+cyc100 promoter</b>. | ||
| + | </p> | ||
| + | <p> | ||
| + | To test the function of PPD sensor part, we used mCherry to report the transcription ability of <b>cyc100 mini promoter</b>. | ||
| + | </p> | ||
| + | <div class="figure"> | ||
| + | <img class="center" src="https://static.igem.org/mediawiki/2013/1/1c/Tsinghua-sensor-4.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 4. Mechanism of PPD sensor | |
| - | + | </p> | |
</div> | </div> | ||
| - | <p>To transfer the system into yeast, we cloned the synthesized part into pRS423, a multi-copy expression vector used in yeast. The pMV-LuxR vector and pRS423 was cut by SpeI、BamHI. Result is shown in Figure | + | |
| - | + | </div> | |
| - | <div class="figure"> | + | |
| + | <div class="section section3"> | ||
| + | <h2>Experiments and Results</h2> | ||
| + | |||
| + | <h3>PPD sensor system constructing</h3> | ||
| + | <div class="figure"> | ||
| + | <img class="center" src="https://static.igem.org/mediawiki/2013/4/47/Tsinghua-sensor-5.png"/> | ||
| + | <p class="legend"> | ||
| + | Figure 5. Overview of PPD sensor constructing | ||
| + | </p> | ||
| + | </div> | ||
| + | <p> | ||
| + | We designed a basic part in our sensor system and synthesized it. The map of the part is shown below. The main part contains triple VP16, Nuclear Location Sequence (NLS), LuxR, pLux and cyc100 mini promoter. | ||
| + | </p> | ||
| + | <div class="figure"> | ||
| + | <img class="center" src="https://static.igem.org/mediawiki/2013/9/9b/Tsinghua-sensor-006.png"/> | ||
| + | <p class="legend"> | ||
| + | Figure 6. Synthesized plasmid containing tVP16-LuxR-Plux-cyc100 mini promoter | ||
| + | </p> | ||
| + | </div> | ||
| + | <p> | ||
| + | To transfer the system into yeast, we cloned the synthesized part into pRS423, a multi-copy expression vector used in yeast. The pMV-LuxR vector and pRS423 was cut by SpeI、BamHI. Result is shown in Figure 7. | ||
| + | </p> | ||
| + | <div class="figure"> | ||
<img class="center" src="https://static.igem.org/mediawiki/2013/5/54/Tsinghua-sensor5.png"/> | <img class="center" src="https://static.igem.org/mediawiki/2013/5/54/Tsinghua-sensor5.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 7. Restriction enzyme cutting result for Luxr_ZHU_MENG and pRS423 | |
| - | + | </p> | |
</div> | </div> | ||
| - | <p>LuxR_ZHU_Meng fragment was ligased with pRS423 fragment. We got the following Clone1 plasmid. | + | <p> |
| - | + | LuxR_ZHU_Meng fragment was ligased with pRS423 fragment. We got the following Clone1 plasmid. | |
| - | <div class="figure"> | + | </p> |
| - | <img class="center" src="https://static.igem.org/mediawiki/2013/f/ | + | <div class="figure"> |
| + | <img class="center" src="https://static.igem.org/mediawiki/2013/f/fb/Tsinghua-sensor-008.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 8. Clone1 | |
| - | + | </p> | |
</div> | </div> | ||
| - | <p>Then we added TEF promoter before LuxR_ZHU_MENG part. As shown in the following picture, pTEF and Clone1 plasmid was digested by SacI and SpeI. Then the two fragments were ligased to construct Clone2 plasmid. | + | <p> |
| - | + | Then we added TEF promoter before LuxR_ZHU_MENG part. As shown in the following picture, pTEF and Clone1 plasmid was digested by SacI and SpeI. Then the two fragments were ligased to construct Clone2 plasmid. | |
| - | <div class="figure"> | + | </p> |
| + | <div class="figure"> | ||
<img class="center" src="https://static.igem.org/mediawiki/2013/8/86/Tsinghua-sensor7.png"/> | <img class="center" src="https://static.igem.org/mediawiki/2013/8/86/Tsinghua-sensor7.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 9. Restriction enzyme cutting result for pTEF and Clone1 | |
| - | + | </p> | |
</div> | </div> | ||
<div class="figure"> | <div class="figure"> | ||
| - | <img class="center" src="https://static.igem.org/mediawiki/2013/ | + | <img class="center" src="https://static.igem.org/mediawiki/2013/4/48/Tsinghua-sensor-0010.png"/> |
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 10. Clone2 | |
| - | + | </p> | |
</div> | </div> | ||
| - | <p>Then, we insert a mCherry after the cyc100 mini promoter as a reporter for the sensor system. We used BamHI and EcoRI to digest the Clone2 and mCherry PCR product. pTF4 was constructed by ligasing these two fragments. | + | <p> |
| - | + | Then, we insert a mCherry after the cyc100 mini promoter as a reporter for the sensor system. We used BamHI and EcoRI to digest the Clone2 and mCherry PCR product. pTF4 was constructed by ligasing these two fragments. | |
| - | <div class="figure"> | + | </p> |
| + | <div class="figure"> | ||
<img class="center" src="https://static.igem.org/mediawiki/2013/a/a4/Tsinghua-sensor9.png"/> | <img class="center" src="https://static.igem.org/mediawiki/2013/a/a4/Tsinghua-sensor9.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 11. Restriction enzyme cutting result for Clone2 and mCherry | |
| - | + | </p> | |
</div> | </div> | ||
<div class="figure"> | <div class="figure"> | ||
| - | <img class="center" src="https://static.igem.org/mediawiki/2013/ | + | <img class="center" src="https://static.igem.org/mediawiki/2013/f/fc/Tsinghua-sensor-0012.png"/> |
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 12. pTF4 | |
| - | + | </p> | |
</div> | </div> | ||
| - | < | + | <h3>PPD sensor system testing</h3> |
| - | <p>We tested the feasibility of our PPD sensor system.</p> | + | <p> |
| - | <p>At first, we wanted to detect whether LuxR protein was expressed in the yeast. The tVP16- | + | We tested the feasibility of our PPD sensor system. |
| - | + | </p> | |
| - | <div class="figure"> | + | <p> |
| + | At first, we wanted to detect whether LuxR protein was expressed in the yeast. The tVP16-LuxR protein was tagged by flag. We transformed the pTF4 into yeast and cultured for 24h before being lysed by 0.2 M NaOH. The yeast total protein was treated by SDS sample buffer. The samples, as well as the control group, were separated by SDS-PAGE and detected by anti-flag antibody. Result is show below. | ||
| + | </p> | ||
| + | <div class="figure"> | ||
<img class="center" src="https://static.igem.org/mediawiki/2013/4/40/Tsinghua-sensor11.png"/> | <img class="center" src="https://static.igem.org/mediawiki/2013/4/40/Tsinghua-sensor11.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 13. Expression of tVP16-LuxR in yeast | |
| - | + | </p> | |
</div> | </div> | ||
| - | <p>From Figure | + | <p> |
| - | + | From Figure 13, we can know that compared with control group, pTF4 transformed yeast had higher expression level of tVP16-LuxR. However, it seems that there is a weak band in control group. We speculated that it might be caused by non-specific binding of anti-flag first antibody. | |
| - | <p> | + | </p> |
| - | + | <p> | |
| - | + | Then we examine the transcription potency of the PPD sensor. We transformed the pTF4 into yeast. One group was treated with 0.5 μM AHL, while the other not. After 24 h inducing, compared with control group, the AHL inducing group had more mCherry positive cells. The several mCherry cells in the control group might be caused by the leakage of the pLux-cyc100 mini promoter. | |
| - | <div class="figure"> | + | </p> |
| + | <div class="figure"> | ||
<img class="center" src="https://static.igem.org/mediawiki/2013/b/ba/Tsinghua-sensor12.png"/> | <img class="center" src="https://static.igem.org/mediawiki/2013/b/ba/Tsinghua-sensor12.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 14. PPD sensor induced by 0.5 uM AHL | |
| - | + | </p> | |
</div> | </div> | ||
| - | <p>The efficiency of | + | <p> |
| - | + | The efficiency of PPD sensor system had been quantified by virtue of flow cytometry. This time, we set three groups. They were induced by different concentrations of AHL (0 μM, 0.5 μM, 200 μM). The fluorescence intensity of these three groups were tested after three different time period (0.5hr, 16hr and 24hr). The result is shown in Figure 15. | |
| - | <div class="figure"> | + | </p> |
| - | <img class="center" src="https://static.igem.org/mediawiki/2013/ | + | <div class="figure"> |
| + | <img class="center" src="https://static.igem.org/mediawiki/2013/c/ca/Tsinghua-sensor-15.png"/> | ||
<p class="legend"> | <p class="legend"> | ||
| - | + | Figure 15. Flow cytometry analysis of AHL inducing result in different time points | |
| - | + | </p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| + | <p> | ||
| + | From the result, we can conclude that, after AHL inducing, there is a significant increase in mCherry fluorescence intensity with the increase of AHL concentration. In 200 μM group, mCherry fluorescence intensity fold change increase from 0.5hr to 16hr, and peaked in 16hr. However, the fluorescence intensity fold change decrease in 24hr, which, we speculated, might be caused by the side effect of high-concentration AHL to the yeast and the elongated time of incubation. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <div class="section section4"> | ||
| + | <h2>Discussion</h2> | ||
| + | <p> | ||
| + | Although our PPD sensor worked, the design of the system has some problems. | ||
| + | </p> | ||
| + | <p> | ||
| + | We did not add terminator after LuxR gene. So read-through may cause the leakage of mCherry transcription and expression. Also, lacking a terminator will decrease the stability and expression efficiency of LuxR protein. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <div id="references"> | ||
| + | <h2>Reference</h2> | ||
| + | <p> | ||
| + | [1] Simon C. Williams et al. Pseudomonas aeruginosa Autoinducer Enters and Functionsin Mammalian Cells. 2004, JOURNALOF BACTERIOLOGY, Vol.186,No.8 | ||
| + | </p> | ||
| + | <p> | ||
| + | [2] F. Sherman et al. Getting Started with Yeast By Fred Sherman, 2003, Methods Enzymol. 350, 3-41. | ||
| + | </p> | ||
| + | <p> | ||
| + | Sergi Regot Rodríguez de Mier, Systems and synthetic biology studies in Saccharomyces cerevisiae. 2011, Barcelona. | ||
| + | </p> | ||
| + | <p> | ||
| + | [4] Quorum sensing in biofilms: Why bacteria behave the way they do? 2009, Journal of Food Science. 74(1): R24–R37. | ||
| + | </p> | ||
| + | <p> | ||
| + | [5] Avantika Lal, Quorum Sensing : How Bacteria Talk to Each Other, 2009, RESONANCE 866-871. | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <script type="text/javascript"> | ||
| + | setupSectionNavs(); | ||
| + | </script> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
</body></html> | </body></html> | ||
Latest revision as of 01:55, 28 September 2013

PPD Sensor
We created PPD sensor part based on quorum sensing system in yeast. The sensor yeast can sense the AHL secreted by bacteria in surroundings. After AHL inducing, the reporter gene mCherry in sensor system can be expressed successfully.
Overview
We constructed the PPD sensor part by reconstructing bacteria quorum sensing system in yeast.
We confirm that the Lux Receptor in the sensor was expressed.
We realized the inducing regulation of our sensor part by adding AHL to the yeast.

Figure 1. Overview of PPD sensor
Design
Designed PPD sensor part
To achieve the sensing of the AHLs from the pathogenic organisms, we plan to introduce the quorum sensing system to yeast, which has been naturally used by the pathogenic microoganisms themselves for communicating within the colonies. Usually, the quorum sensing system share the similar working mechanisms between different species, the precise procedures are shown as follows:

Figure 2. Principle of quorum sensing system
Reconstruction the PPD system
Considered the difference between prokaryotic and eukaryotic system, the prokaryotic quorum sensing system needs to be reconstructed in a “eukaryotic way” when introduced into yeast. As the quorum sensing systems in different species share the similar mechanism, we typically reconstructed the LuxR system.

Figure 3. Reconstruction of quorum sensing system in yeast
For the reconstruction of LuxR, we add three times repeat of VP16 region, as well as a region of NLS. The 3 times repeated sequence of VP16 is function as activation domain (AD) to link the RNA POL II, the Nuclear Localization Sequence (NLS) is aimed to import the LuxR to the nuclear and thus enhance the transcriptional activating potency.
For the reconstruction of Lux Promoter, we add cyc100 mini promoter (the TATA Box), the function of which is to recruit RNA POL II to DNA region.
These modifications will lead to the effect that when AHL is introduced into the cell, LuxR will bind with AHL and then together bind with Lux promoter, the VP16 domain of the complex will then bind with POL II, which bind to cyc100 mini promoter and facilitate the gene expression downstream of plux+cyc100 promoter.
To test the function of PPD sensor part, we used mCherry to report the transcription ability of cyc100 mini promoter.

Figure 4. Mechanism of PPD sensor
Experiments and Results
PPD sensor system constructing

Figure 5. Overview of PPD sensor constructing
We designed a basic part in our sensor system and synthesized it. The map of the part is shown below. The main part contains triple VP16, Nuclear Location Sequence (NLS), LuxR, pLux and cyc100 mini promoter.
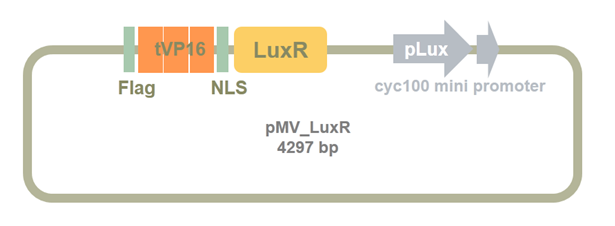
Figure 6. Synthesized plasmid containing tVP16-LuxR-Plux-cyc100 mini promoter
To transfer the system into yeast, we cloned the synthesized part into pRS423, a multi-copy expression vector used in yeast. The pMV-LuxR vector and pRS423 was cut by SpeI、BamHI. Result is shown in Figure 7.

Figure 7. Restriction enzyme cutting result for Luxr_ZHU_MENG and pRS423
LuxR_ZHU_Meng fragment was ligased with pRS423 fragment. We got the following Clone1 plasmid.

Figure 8. Clone1
Then we added TEF promoter before LuxR_ZHU_MENG part. As shown in the following picture, pTEF and Clone1 plasmid was digested by SacI and SpeI. Then the two fragments were ligased to construct Clone2 plasmid.

Figure 9. Restriction enzyme cutting result for pTEF and Clone1
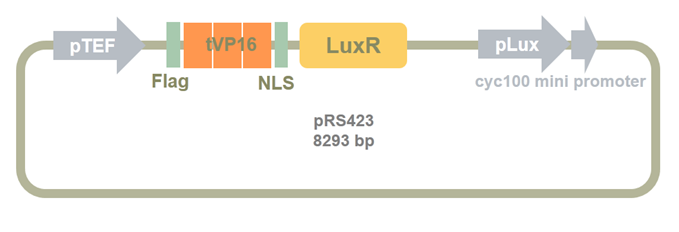
Figure 10. Clone2
Then, we insert a mCherry after the cyc100 mini promoter as a reporter for the sensor system. We used BamHI and EcoRI to digest the Clone2 and mCherry PCR product. pTF4 was constructed by ligasing these two fragments.

Figure 11. Restriction enzyme cutting result for Clone2 and mCherry
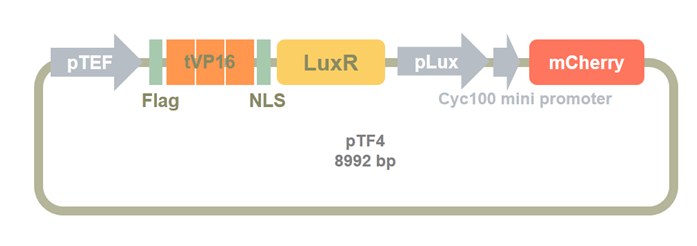
Figure 12. pTF4
PPD sensor system testing
We tested the feasibility of our PPD sensor system.
At first, we wanted to detect whether LuxR protein was expressed in the yeast. The tVP16-LuxR protein was tagged by flag. We transformed the pTF4 into yeast and cultured for 24h before being lysed by 0.2 M NaOH. The yeast total protein was treated by SDS sample buffer. The samples, as well as the control group, were separated by SDS-PAGE and detected by anti-flag antibody. Result is show below.

Figure 13. Expression of tVP16-LuxR in yeast
From Figure 13, we can know that compared with control group, pTF4 transformed yeast had higher expression level of tVP16-LuxR. However, it seems that there is a weak band in control group. We speculated that it might be caused by non-specific binding of anti-flag first antibody.
Then we examine the transcription potency of the PPD sensor. We transformed the pTF4 into yeast. One group was treated with 0.5 μM AHL, while the other not. After 24 h inducing, compared with control group, the AHL inducing group had more mCherry positive cells. The several mCherry cells in the control group might be caused by the leakage of the pLux-cyc100 mini promoter.

Figure 14. PPD sensor induced by 0.5 uM AHL
The efficiency of PPD sensor system had been quantified by virtue of flow cytometry. This time, we set three groups. They were induced by different concentrations of AHL (0 μM, 0.5 μM, 200 μM). The fluorescence intensity of these three groups were tested after three different time period (0.5hr, 16hr and 24hr). The result is shown in Figure 15.

Figure 15. Flow cytometry analysis of AHL inducing result in different time points
From the result, we can conclude that, after AHL inducing, there is a significant increase in mCherry fluorescence intensity with the increase of AHL concentration. In 200 μM group, mCherry fluorescence intensity fold change increase from 0.5hr to 16hr, and peaked in 16hr. However, the fluorescence intensity fold change decrease in 24hr, which, we speculated, might be caused by the side effect of high-concentration AHL to the yeast and the elongated time of incubation.
Discussion
Although our PPD sensor worked, the design of the system has some problems.
We did not add terminator after LuxR gene. So read-through may cause the leakage of mCherry transcription and expression. Also, lacking a terminator will decrease the stability and expression efficiency of LuxR protein.
Reference
[1] Simon C. Williams et al. Pseudomonas aeruginosa Autoinducer Enters and Functionsin Mammalian Cells. 2004, JOURNALOF BACTERIOLOGY, Vol.186,No.8
[2] F. Sherman et al. Getting Started with Yeast By Fred Sherman, 2003, Methods Enzymol. 350, 3-41.
Sergi Regot Rodríguez de Mier, Systems and synthetic biology studies in Saccharomyces cerevisiae. 2011, Barcelona.
[4] Quorum sensing in biofilms: Why bacteria behave the way they do? 2009, Journal of Food Science. 74(1): R24–R37.
[5] Avantika Lal, Quorum Sensing : How Bacteria Talk to Each Other, 2009, RESONANCE 866-871.
 "
"
