Team:NCTU Formosa/project
From 2013.igem.org
Mike821005 (Talk | contribs) (→Introduction) |
(→i-Culture) |
||
| (93 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<div class="li"><div class="card"> | <div class="li"><div class="card"> | ||
===Introduction=== | ===Introduction=== | ||
| - | <p>The majority of the previous iGEM projects focused on expressing certain genes to achieve a specific task such as sensing a certain phenomenon. This year, we took a step forward in exploring the potential of '' | + | <p>The majority of the previous iGEM projects focused on expressing certain genes to achieve a specific task such as sensing a certain phenomenon. This year, we took a step forward in exploring the potential of ''Escherichia coli'', thus building a system that is capable of '''regulating multiple genes'''. The system, so-called E.colightuner, is built by three regulated-systems including '''light regulated-system''', '''temperature regulated-system''', and '''sRNA regulated-system'''. We used noninvasive factors such as red light and temperature to create different conditions under which ''E. coli'' can express different genes. We also introduced a new sRNA regulated-system that hasn't been widely applied in iGEM before. </p> |
| - | <p>E.colightuner is just like a machine that can process different tasks depending on the commands given. In this case, the gene expression of E.colightuner is the task and its environmental condition is the command. By changing | + | <p>E.colightuner is just like a machine that can process different tasks depending on the commands given. In this case, the gene expression of E.colightuner is the task and its environmental condition is the command. By changing the environmental condition, E.colightuner can express different genes. The advantage of building such a machine is to increase the convenience in expressing multiple genes. Instead of expressing different genes with different bacteria, we can express the genes with E.colightuner. This may ultimately lower the cost in expressing different genes.</p> |
| - | + | ||
<p>We tend to apply E.colightuner to agriculture. By expressing different hormones required by different stages of plant growth, E.colightuner can aid plant growth. With that said, we built a device that can flower the desired hormones to plants. This device can be applied to any plantation, and it is simple to manipulate. With more efforts, we believe that E.colightuner can raise the potential values of ''E.coli''.</p> | <p>We tend to apply E.colightuner to agriculture. By expressing different hormones required by different stages of plant growth, E.colightuner can aid plant growth. With that said, we built a device that can flower the desired hormones to plants. This device can be applied to any plantation, and it is simple to manipulate. With more efforts, we believe that E.colightuner can raise the potential values of ''E.coli''.</p> | ||
| - | + | [[File:Nctu_formosa_introduction1.jpg|center|600px|Figure 1. We attempt to construct a regulated system that can express different genes under different conditions.]] | |
| - | [[File:Nctu_formosa_introduction1.jpg|center|600px| | + | |
</div></div> | </div></div> | ||
| Line 50: | Line 48: | ||
Light is an easily accessible energy that can be applied to various fields. Of course, it can be applied to synthetic biology as well. Bacteria can sense light of a defined wavelength through a small light-absorbing ligand called chromophore. By employing this chromophore, light can be used to trigger a certain reaction and even regulate gene expressions. | Light is an easily accessible energy that can be applied to various fields. Of course, it can be applied to synthetic biology as well. Bacteria can sense light of a defined wavelength through a small light-absorbing ligand called chromophore. By employing this chromophore, light can be used to trigger a certain reaction and even regulate gene expressions. | ||
| - | In E.colightuner, light serves as an activator that can regulate the function of a promoter. Since red light can be easily turned on and off, the activation and repression of the promoter can also be easily achieved. Substrate activators, on the other hand, are harder to manipulate. Substrates can activate their corresponding promoters efficiently, but they might be hard to remove,making it difficult to repress the promoters when desired. | + | In E.colightuner, light serves as an activator that can regulate the function of a promoter. Since red light can be easily turned on and off, the activation and repression of the promoter can also be easily achieved. Substrate activators, on the other hand, are harder to manipulate. Substrates can activate their corresponding promoters efficiently, but they might be hard to remove, making it difficult to repress the promoters when desired. |
| - | <p>In order to make '' | + | <p>In order to make ''E. coli,'' sense and respond to light, we installed a '''"two component system" (cph8)''' that can respond to '''red light'''. </p> |
| - | <p>The chimera | + | <p>The chimera cph8 is consisted of a phytochrome cph1 and a histidine kinase domain, envz-ompR, that includes a response regulator. cph1 is the first member of the plant photoreceptor family that has been identified in bacteria. |
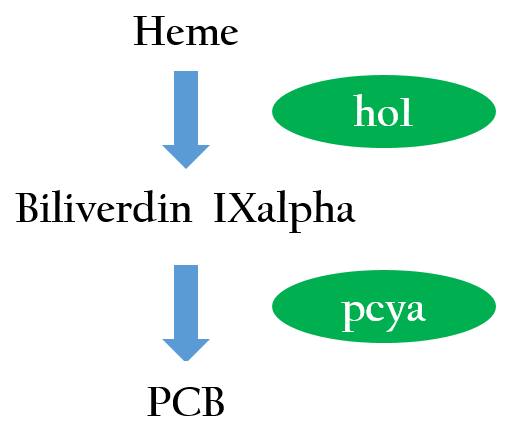
| - | The functional expression of a phytochrome domain( | + | The functional expression of a phytochrome domain (cph1) in ''E. coli'' requires the biosynthesis of the respective bilin chromophore '''PCB'''. Two phycocyanobilin(PCB)-biosynthesis genes (ho1<sup>1</sup> and pcya<sup>2</sup>) from Synechocystis encoding a heme oxygenase and bilin reductase, respectively, is essential to convert heme into PCB. Figure 2 depicts the phycocyanobilin-biosynthesis pathway.<sup>3</sup> With the biosynthesis of PCB, cph1 serve as a red-light absorbing chromophore that is inactivated under red light and activated without red light . <sup>4,5</sup></p> |
| - | [[File:Nctu_formosa_PCBsynthe.jpg|center|300px| | + | [[File:Nctu_formosa_PCBsynthe.jpg|center|300px|Figure 2. Phycocyanobilin is the part of photoreceptor that responds to light, and it is not naturally produced in ''E.coli'',so we introduced two phycocyanobilin-biosynthesis genes (ho1 and pcya) from Synechocystis that convert heme into phycocyanobilin.]] |
| - | <p> | + | <p>envZ-ompR, a dimeric osmosensor, is a multidomain transmembrane protein and one of the best characterized two-component histidine kinases from ''E.coli''. Besides the periplasmic receptor domain that is flanked by two transmembrane helices, it possesses a C-terminal 228-residue histidine kinase domain that is located in the cytoplasm<sup>6</sup>. |
| - | Upon changes of extracellular osmolarity, | + | Upon changes of extracellular osmolarity, envZ specifically phosphorylates its cognate response regulator ompR, which, in turn, regulates the '''P<sub>ompc</sub>''' <sup>7</sup>.</p> |
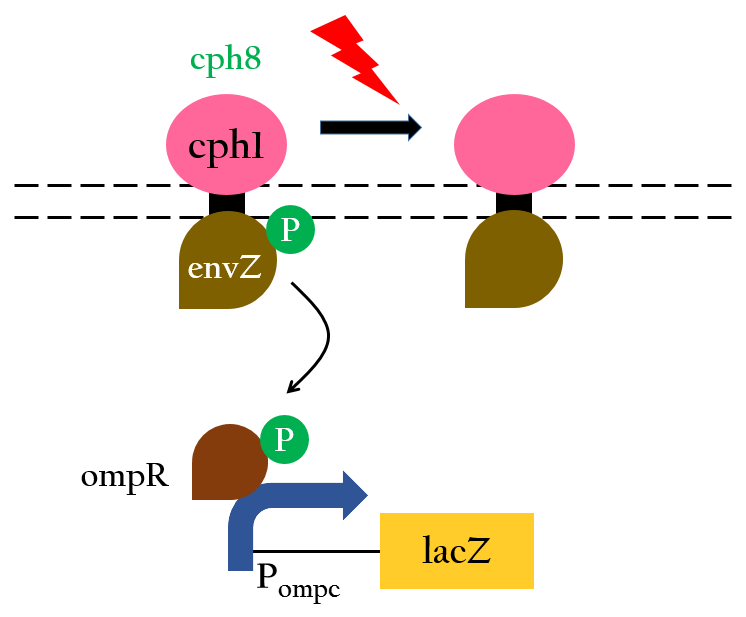
| - | <p>With the two components just mentioned, | + | <p>With the two components just mentioned, cph8 can serve as a photoreceptor that regulates gene expression through P<sub>ompc</sub>. Without red light, cph1 is activated and it enables envZ-ompR to autophosphorylate which in turn activates P<sub>ompc</sub>. Under the exposure of red light, however, cph1 is deactivated, inhibiting the autophosphorylation, thus turning off gene expression. This mechanism is depicted in Figure 3<sup>8</sup> and the experimental data that support this mechanism is shown in Figure 4.</p> |
| - | [[File:Nctu_formosa_cph8mechnism.jpg|center|450px| | + | [[File:Nctu_formosa_cph8mechnism.jpg|center|450px|Figure 3. The light receptor cph8 is composed of cph1(pink) and envZ-ompR(maroon).]] |
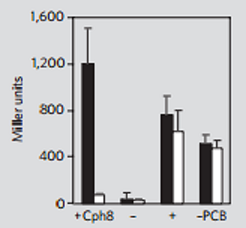
| - | [[File:NCTU_PCBfunction.png|center| | + | [[File:NCTU_PCBfunction.png|center|Figure 4. Miller assay (lacZ expression)showing that cph8 is active in the dark (black bars) in the presence of PCB and inactive in the light (white bars). There is no light-dependent activity in the absence of cph8 (-) and there is constitutive activity when only the histidine kinase domain of envZ is expressed (+) , or when the PCB metabolic pathway is not included (-PCB). from Levskaya, A. et al .(2005). Engineering Escherichia coli to see light. Nature, 438(7067), 442. ]] |
====Design of Red promoter==== | ====Design of Red promoter==== | ||
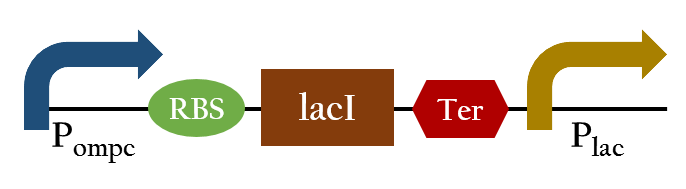
| - | We designed a new biobrick, P<sub>red</sub>, to '''positively''' regulate gene expression using red light. Compared to the negative regulation provided by | + | We designed a new biobrick, P<sub>red</sub>, to '''positively''' regulate gene expression using red light. Compared to the negative regulation provided by cph8, positive regulation is more direct and convenient to operate. In addition, cph8 relies on P<sub>ompc</sub> to express downstream gene, but our design relies on a stronger promoter, P<sub>lac</sub>, that can amplify gene expression, further making sure that the level of expression can meet our needs. |
| - | [[File:Nctu_formosa_pred.png|center|500px| | + | [[File:Nctu_formosa_pred.png|center|500px|Figure 5. The P<sub>red</sub> promoter.]] |
| - | In the presence of red light, P<sub>ompc</sub> is shut down and lacI cannot be expressed to repress P<sub>lac</sub>. As a result, P<sub>lac</sub> is activated and all the downstream genes can be transcribed. In the absence of red light, whatsoever, P<sub>ompc</sub> is activated as autophosphorylation is allowed. LacI can | + | In the presence of red light, P<sub>ompc</sub> is shut down and lacI cannot be expressed to repress P<sub>lac</sub>. As a result, P<sub>lac</sub> is activated and all the downstream genes can be transcribed. In the absence of red light, whatsoever, P<sub>ompc</sub> is activated as autophosphorylation is allowed. LacI can be expressed to repress P<sub>lac</sub> and the genes downstream cannot be expressed, consequently. By using P<sub>red</sub>, we can achieve positive control using red light. |
======Reference====== | ======Reference====== | ||
| Line 102: | Line 100: | ||
It is not a dream anymore that "making the impossible possible" with the increase of modern technology. It is also a serious problem that what human has brought to Earth along with our progressing civilization. Drastic environmental changes are inevitable due to the fact of massive developing industries. Temperature changing are one of the problems we need to confront. | It is not a dream anymore that "making the impossible possible" with the increase of modern technology. It is also a serious problem that what human has brought to Earth along with our progressing civilization. Drastic environmental changes are inevitable due to the fact of massive developing industries. Temperature changing are one of the problems we need to confront. | ||
| - | + | Cells are constantly subjected to changing environmental conditions. Comparing with chemical substances, temperature is more accessible, convenient and lower costing. Moreover, temperature is also less burden to the organisms. Therefore, few organisms use temperature as a regulating factor. A mechanism found in different organisms that makes the cell respond to thermal changes, for example, is the '''RNA thermometer'''. | |
| - | An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. The expression of heat-shock, cold-shock and some virulence genes are coordinated in response to temperature changes.There are several systems suggested in references that are based on RNA secondary structure. This structural transition can then expose or occlude important regions of RNA such as a'''ribosome binding site''', which then affects the translation rate of a nearby protein-coding gene, so we can say that the RNA thermometers are just like riboswitches. | + | An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. The expression of heat-shock, cold-shock and some virulence genes are coordinated in response to temperature changes. There are several systems suggested in references that are based on RNA secondary structure. This structural transition can then expose or occlude important regions of RNA such as a '''ribosome binding site''', which then affects the translation rate of a nearby protein-coding gene, so we can say that the RNA thermometers are just like riboswitches. |
Apart from protein-mediated transcriptional control mechanisms, translational control by RNA thermometers is also a widely used regulatory strategy. | Apart from protein-mediated transcriptional control mechanisms, translational control by RNA thermometers is also a widely used regulatory strategy. | ||
| Line 111: | Line 109: | ||
RNA thermometers are identified in the 5' UTR of messenger RNA, upstream of a protein-coding gene. | RNA thermometers are identified in the 5' UTR of messenger RNA, upstream of a protein-coding gene. | ||
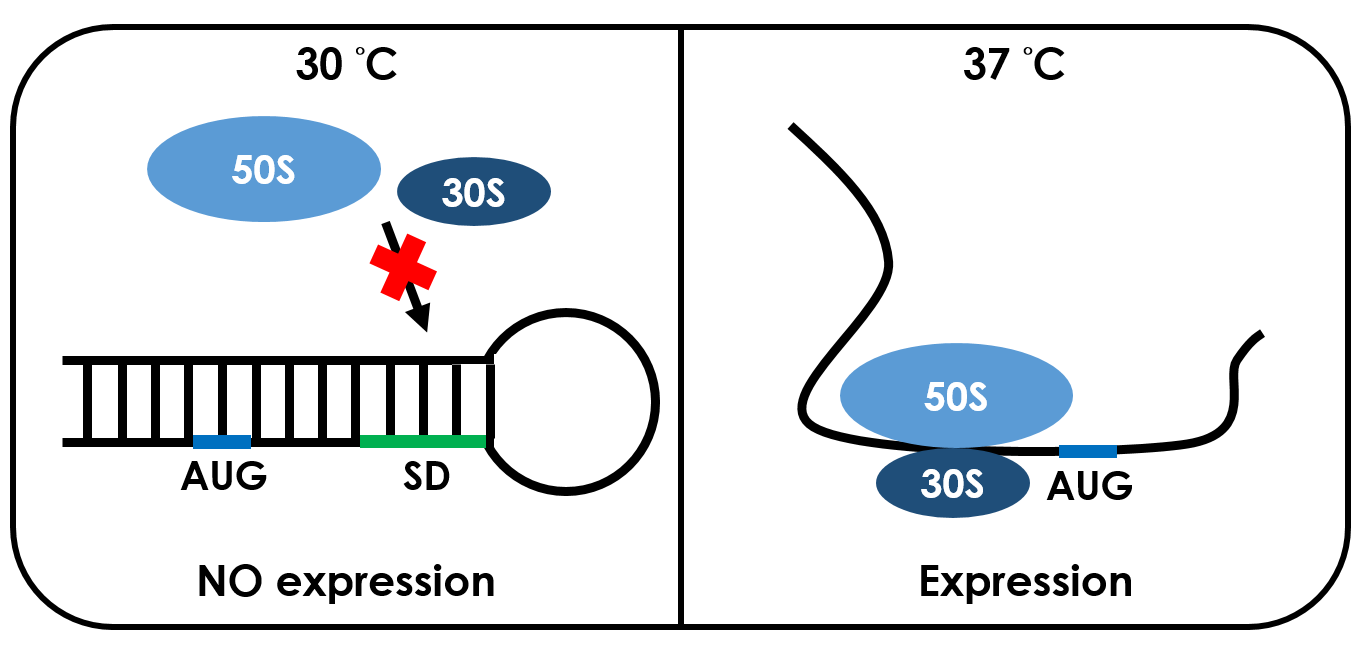
| - | Unlike normal RBS, the ''' | + | Unlike normal RBS, the '''37 °C RBS''' has a unique or rather stable '''hairpin''' structure which shaped like a lock. If the temperature drops below 37 °C, the base-pairing neucleotides on the RNA will form a stable secondary hairpin structure, occluding the '''Shine-Dalgarno sequence''' (SD sequence) thus disabling the ribosome to bind. The base-pairing of this RNA region will block the expression of the protein encoded within. Since the structure is sustained by base pairing, heat can be employed to break the hydrogen bonds. So when it reaches 37 °C, the bonds would be broken and the hairpin would be unfolded, causing the SD sequence to be exposed and permitting the binding of the ribosome, which then starts the translation. This means that by raising temperature to a certain threshold, the 37 °C RBS can function as a normal RBS. In this way, gene expression can be regulated on the RNA level by temperature. |
| - | [[File:NCTU_RNA1.jpg|center|600px| | + | [[File:NCTU_RNA1.jpg|center|600px|Figure 6. Top left, the RNA forms stable base-pairs on the SD sequence, disabling the ribosome to bind. (SD sequence is the polypurine sequence AGGAGG centered about 10 bp before the AUG initiation codon on bacterial mRNA.) It is now in the off-state. The base-pairing of this RNA region will block the expression of the protein encoded behind it. On the other hand, top right, the highlighted SD sequence becomes exposed, allowing the binding of the 30S ribosomal subunit. Translation can now initiate, because the ribosome is able to bind to the Shine Dalgarno region. The system is now in the on-state. |
]] | ]] | ||
====Design==== | ====Design==== | ||
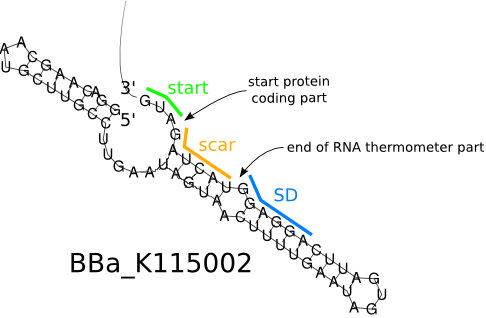
| - | The specific ribosome binding site (RBS) [http://parts.igem.org/Part:BBa_K115002 BBa_ K115002] is a great RNA thermometer. It has high translation activity at high temperature(> | + | The specific ribosome binding site (RBS) [http://parts.igem.org/Part:BBa_K115002 BBa_ K115002] is a great RNA thermometer. It has high translation activity at high temperature (> 37 °C) and low translation activity at room temperature. It was designed for the temperature-dependent genetic circuit in ''E. coli'', with a green fluorescent protein (GFP) used as the reporter protein. |
| - | Originally, it is in the 5'-UTR of the Salmonella ''agsA'' gene, which codes for a small heat shock protein that can be used for temperature sensitive post-transcriptional regulation, initiating translation at | + | Originally, it is in the 5'-UTR of the Salmonella ''agsA'' gene, which codes for a small heat shock protein that can be used for temperature sensitive post-transcriptional regulation, initiating translation at 37 °C. Transcription of the ''agsA'' gene is controlled by the alternative sigma factor σ32. Additional translational control depends on a stretch of four uridines that pair with the SD sequence, so they are named "FourU". They repress translation of this protein by base-pairing with the Shine-Dalgardo sequence of the mRNA. This prevents ribosomes from binding the start codon. |
| - | In our project, we use the | + | In our project, we use the 37 °C RBS in both red light induced circuit and dark response circuit and it is related to the production of GFP and BFP. We use this 37 °C RBS to design our project because we want to use different conditions to produce different substances. Therefore, the characteristic and sensitivity of 37 °C RBS perfectly fits our purpose which we want to regulate our system through temperature. |
| - | [[File:NCTU_BBa_K115002.png|center|800px| | + | [[File:NCTU_BBa_K115002.png|center|800px|Figure 7. The image shows the secondary structure of RNA thermometer as predicted by RNA fold. Notice that the 3' end that includes the scar and the start codon is folded up and not extended. The blue region is the Shine Dalgarno sequence that serves as the ribosome binding site. The blue nucleotides had to be changed in order to get the desired secondary structure after introducing of the scar. ]] |
| Line 144: | Line 142: | ||
<p>Base pairing offers a powerful way for one RNA to control the activity of another. Both prokaryotes and eukaryotes have many cases which a single-stranded RNA base pairs with a complementary region of an mRNA. As a result, it prevents expression of the mRNA.</p> | <p>Base pairing offers a powerful way for one RNA to control the activity of another. Both prokaryotes and eukaryotes have many cases which a single-stranded RNA base pairs with a complementary region of an mRNA. As a result, it prevents expression of the mRNA.</p> | ||
| - | <p>'''RNA interference(RNAi)''', also called post transcriptional gene silencing, is a process in which RNA molecules inhibit gene expression by destroying specific mRNA molecules. In 2006, Andrew Fire and Craig C.Mello shared the Nobel Prize in Physiology or Medicine because of their study on RNA interference<sup>11</sup>. This powerful gene silencing tool in eukaryotes has been used in many research. As their counterparts in bacteria, '''small non-coding RNAs(sRNAs)''' are important regulatory roles.</p> | + | <p>'''RNA interference (RNAi)''', also called post transcriptional gene silencing, is a process in which RNA molecules inhibit gene expression by destroying specific mRNA molecules. In 2006, Andrew Fire and Craig C.Mello shared the Nobel Prize in Physiology or Medicine because of their study on RNA interference<sup>11</sup>. This powerful gene silencing tool in eukaryotes has been used in many research. As their counterparts in bacteria, '''small non-coding RNAs (sRNAs)''' are important regulatory roles.</p> |
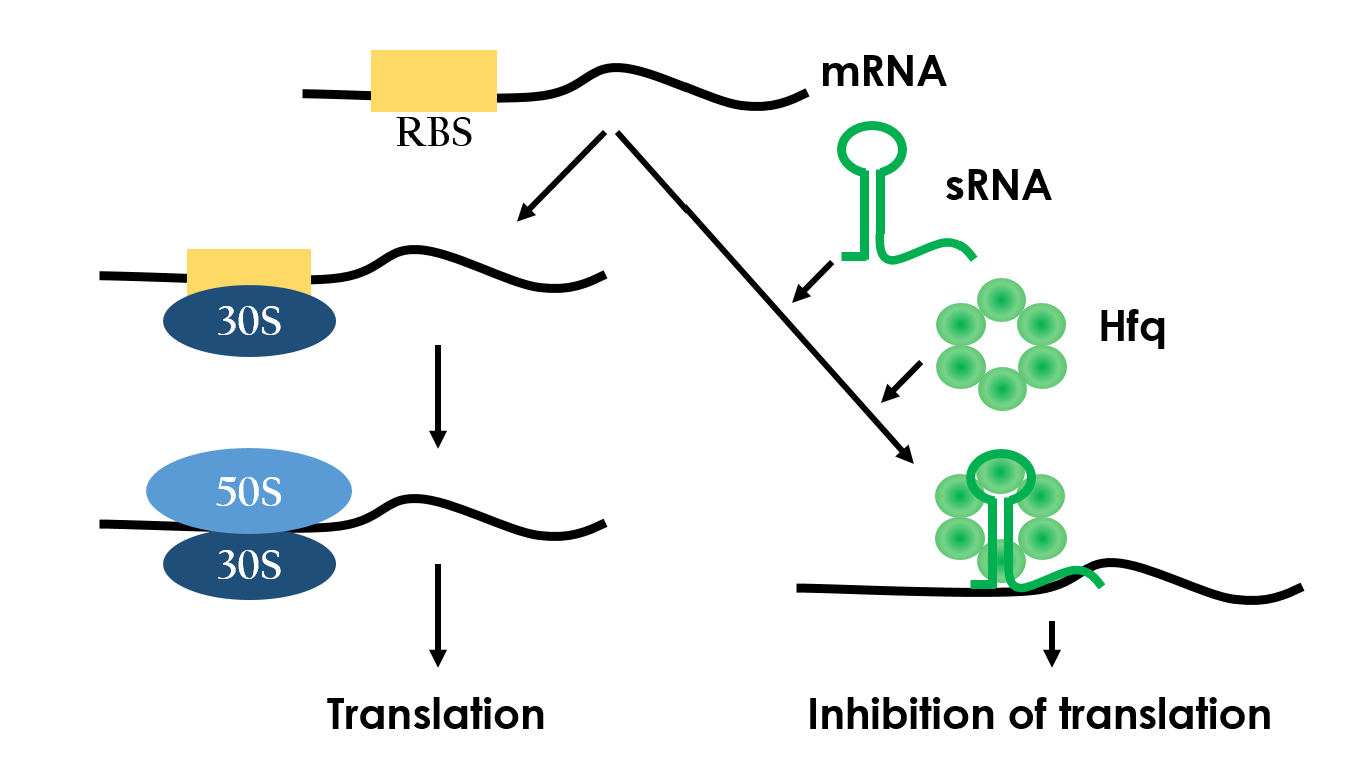
| - | <p>Small RNAs(sRNAs) have become increasingly significant in playing the role of bacterial gene regulation. Most sRNAs interact with the targeted mRNAs by imperfect base pairing, reducing the translation efficiency and recruiting chaperones such as Hfq for translation termination. The sRNAs would bind to the target with its hairpin-like structure which wound by some of it's own sequences, and the chaperons would stuck between the hairpins in order to protect the sRNA-mRNA combination from degrading. In the end, the sRNAs regulate gene expression by forestalling translation. Since this regulated-system has already been employed in vivo, it is necessary to design an artificial sRNA that targets specifically to a desired genes, so that we can prevent the sRNA from affecting other undesired genes.</p> | + | <p>Small RNAs (sRNAs) have become increasingly significant in playing the role of bacterial gene regulation. Most sRNAs interact with the targeted mRNAs by imperfect base pairing, reducing the translation efficiency and recruiting chaperones such as Hfq for translation termination. The sRNAs would bind to the target with its hairpin-like structure which wound by some of it's own sequences, and the chaperons would stuck between the hairpins in order to protect the sRNA-mRNA combination from degrading. In the end, the sRNAs regulate gene expression by forestalling translation. Since this regulated-system has already been employed in vivo, it is necessary to design an artificial sRNA that targets specifically to a desired genes, so that we can prevent the sRNA from affecting other undesired genes.</p> |
| - | <p>Unlike the regulated-system popular used in iGEM projects(e.g. | + | <p>Unlike the regulated-system popular used in iGEM projects (e.g. P<sub>tet</sub> & ''tetR'', P<sub>lux</sub> & ''LuxR'', etc.), sRNA regulated-system seems '''more efficient'''. Because the inhibition works under RNA level, which means the energy wasted in ''E. coli'' is less than other system (producing proteins to regulate).</p> |
Figure 8 depicts how the small RNA chaperone, Hfq, associates with the small RNA and represses a target mRNA.<sup>12</sup> | Figure 8 depicts how the small RNA chaperone, Hfq, associates with the small RNA and represses a target mRNA.<sup>12</sup> | ||
| - | [[File:NCTU_sRNA_Introduction.png|center|600px| | + | [[File:NCTU_sRNA_Introduction.png|center|600px|Figure 8. <br>Hfq and a small RNA may sequester the ribosome-binding site of a target mRNA, thus blocking binding of the 30S and 50S ribosomal subunits and repressing translation.]] |
====Mechanism==== | ====Mechanism==== | ||
<p>How do small RNAs regulate gene expression? | <p>How do small RNAs regulate gene expression? | ||
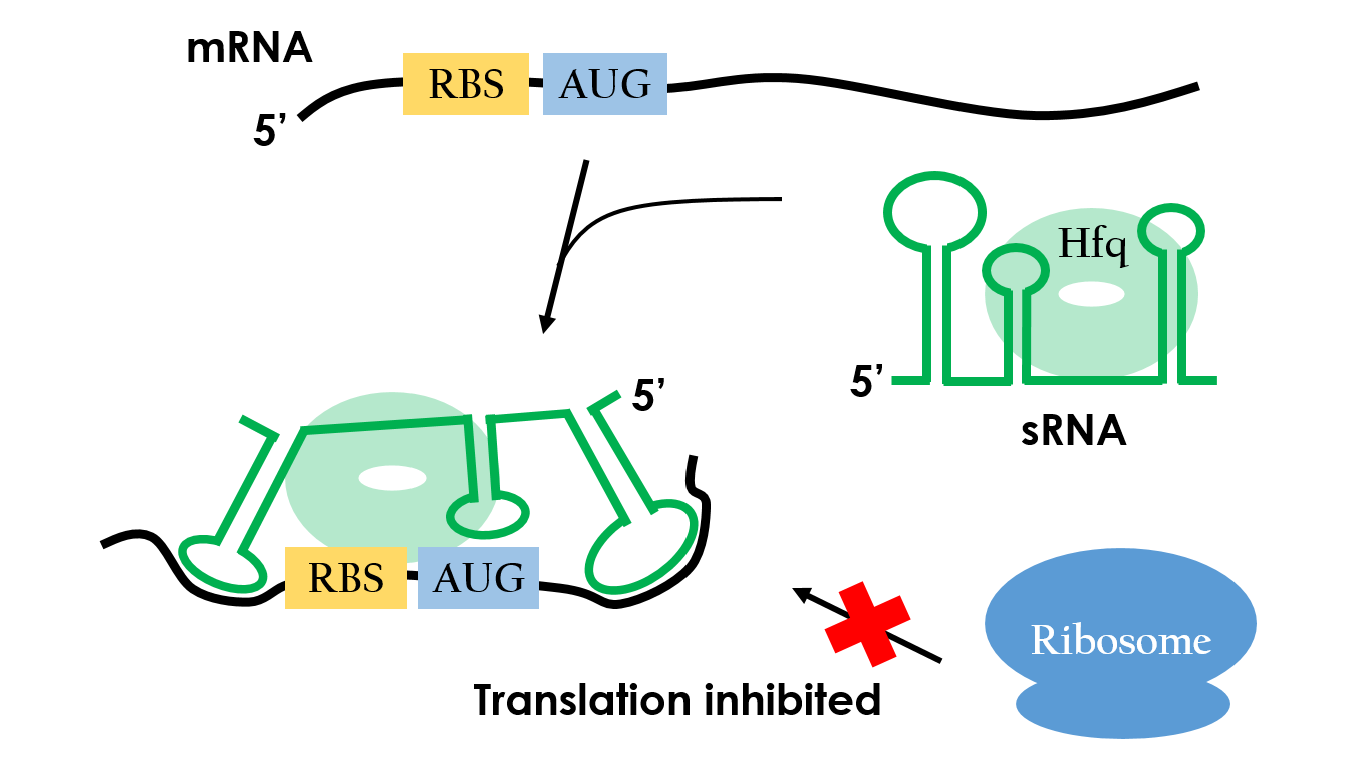
| - | Figure 9 | + | Figure 9 shows the mechanism of repressing one target mRNA. |
The small RNA has three stem-loop double stranded RNA structures, | The small RNA has three stem-loop double stranded RNA structures, | ||
and the loop which is closest to the 3’ terminus is complementary | and the loop which is closest to the 3’ terminus is complementary | ||
| Line 164: | Line 162: | ||
| - | [[File:NCTU FORMOSA WEB WIKI sRNA inhibition.JPG|center|600px| | + | [[File:NCTU FORMOSA WEB WIKI sRNA inhibition.JPG|center|600px|Figure 9. <br>sRNA-Hfq complex specifically binds on target gene, forming a blockade of ribosome binding. Therefore, the translation is inhibited.]] |
====Design==== | ====Design==== | ||
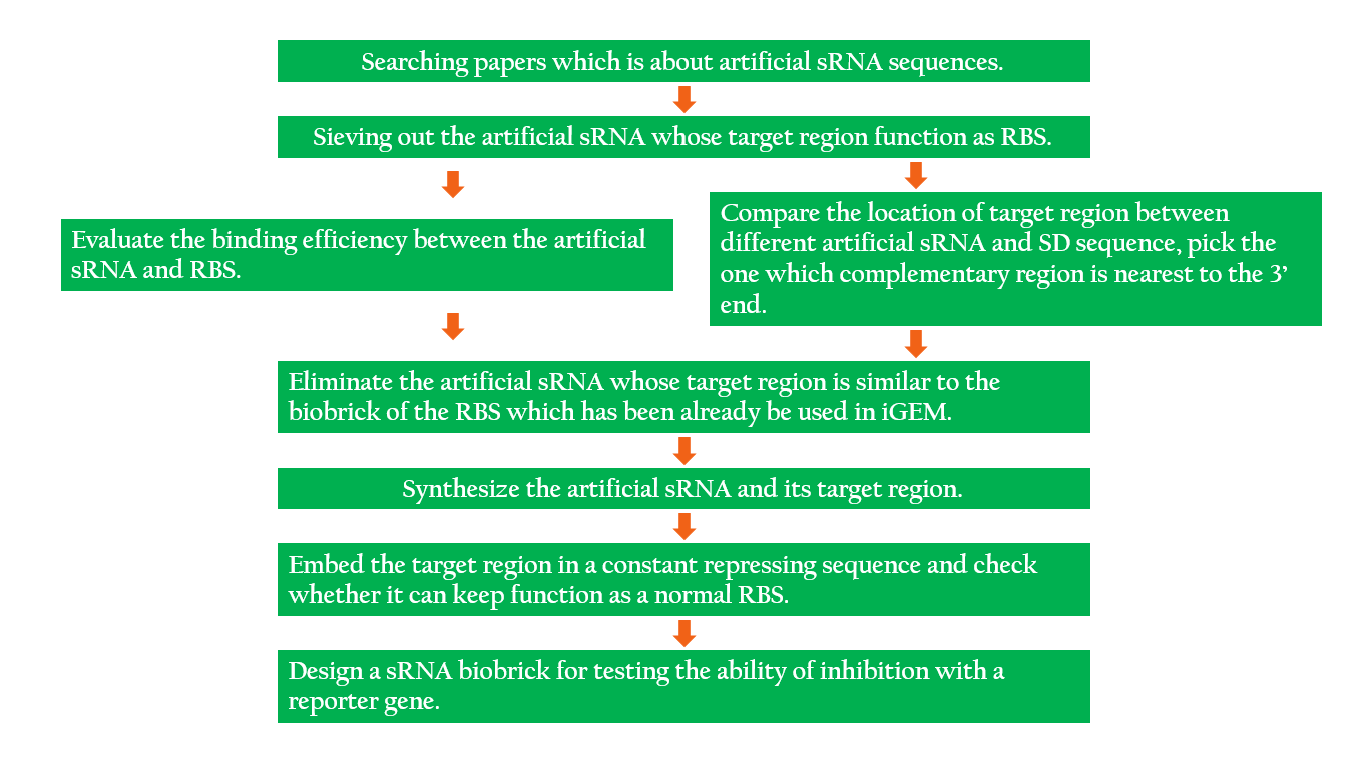
| - | + | The following is the process of our idea in designing the sRNA. | |
| - | [[File:NCTU_sRNA_Design.png|900px|center| | + | [[File:NCTU_sRNA_Design.png|900px|center|Figure 10. Design process.]] |
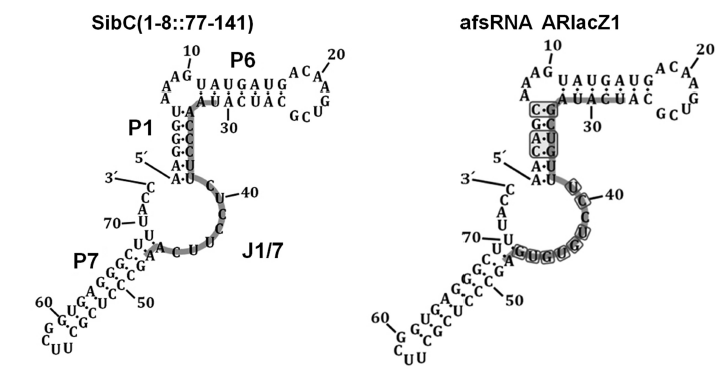
| - | We have designed two artificial sRNAs, each modified from different paper. The sRNA-1 is based on afsRNA ARlacZ1,which is created by Hongmarn Park and his colleagues.<sup>15</sup> ''SibC'' is a small RNA sequence which is already found in ''E.coli''. Hongmarn Park and his colleagues changed few base pairs of ''SibC'' to make ARlacZ1, purposed to make the secondary structure more stable. After that, they tried to change 9 different locations of the target-recognition sequence to test the effect on gene silencing. Our design is to modify from the RNA sequence which has the best efficiency: ARlacZ7. In accordance with afsRNA ARlacZ7, we changed the target-recognition sequence to finally complete our sRNA-1. | + | We have designed two artificial sRNAs, each modified from different paper. The sRNA-1 is based on afsRNA ARlacZ1, which is created by Hongmarn Park and his colleagues.<sup>15</sup> ''SibC'' is a small RNA sequence which is already found in ''E.coli''. Hongmarn Park and his colleagues changed few base pairs of ''SibC'' to make ARlacZ1, purposed to make the secondary structure more stable. After that, they tried to change 9 different locations of the target-recognition sequence to test the effect on gene silencing. Our design is to modify from the RNA sequence which has the best efficiency : ARlacZ7. In accordance with afsRNA ARlacZ7, we changed the target-recognition sequence to finally complete our sRNA-1. |
| - | [[File:Nctu_formosa_srna_design.png|600px|center| | + | [[File:Nctu_formosa_srna_design.png|600px|center|Figure 11. Secondary structure models of SibC and afsRNA ARlacZ1. The replaced bases in ARlacZ1 are boxed. From ''Hongmarn Park, Geunu Bak, Sun Chang Kim & Younghoon Lee.(2013). "Exploring sRNA-mediated gene silencing mechanisms using artificial small RNAs derived from a natural RNA scaffold in Escherichia coli". Nucleic Acids Research,Vol. 41, No. 6, 3787-3804 DOI:10.1093'']] |
| - | The other sRNA sequence we produced is called sRNA-2. It is employed in this project | + | The other sRNA sequence we produced is called sRNA-2. It is employed in this project selected from a library of artificial sRNA that was constructed by fusing a randomized antisense domain of Spot42, the scaffold that is known for recruiting the RNA chaperons. The sRNA we picked contains a consensus sequence, 5’-CCCUC-3’, which can base pair with the SD sequence due to complementary binding. This sRNA, as expected, effectively regulates gene expression by reducing translation efficiency and recruiting Hfq. In addition, this sRNA shows high specificity against its targeted gene, ''OmpF'', as it doesn't hold significant activity against other genes from ''E. coli'' genome (Sharma and others, 2011). |
| - | [[File:Nctu_srna2design.png#file|center|1000px| | + | [[File:Nctu_srna2design.png#file|center|1000px|Figure 12. Artificial sRNA based on the Spot42 sRNA scaffold (yellow box). The bases (red) is |
| - | target-recognition region from Vandana Sharma, Asami Yamamura & Yohei Yokobayashi.(2011). "Engineering Artificial Small RNAs for Conditional Gene Silencing in E. coli". ACS Synthetic Biology.]] | + | target-recognition region from Vandana Sharma, Asami Yamamura & Yohei Yokobayashi. (2011) . "Engineering Artificial Small RNAs for Conditional Gene Silencing in E. coli". ACS Synthetic Biology.]] |
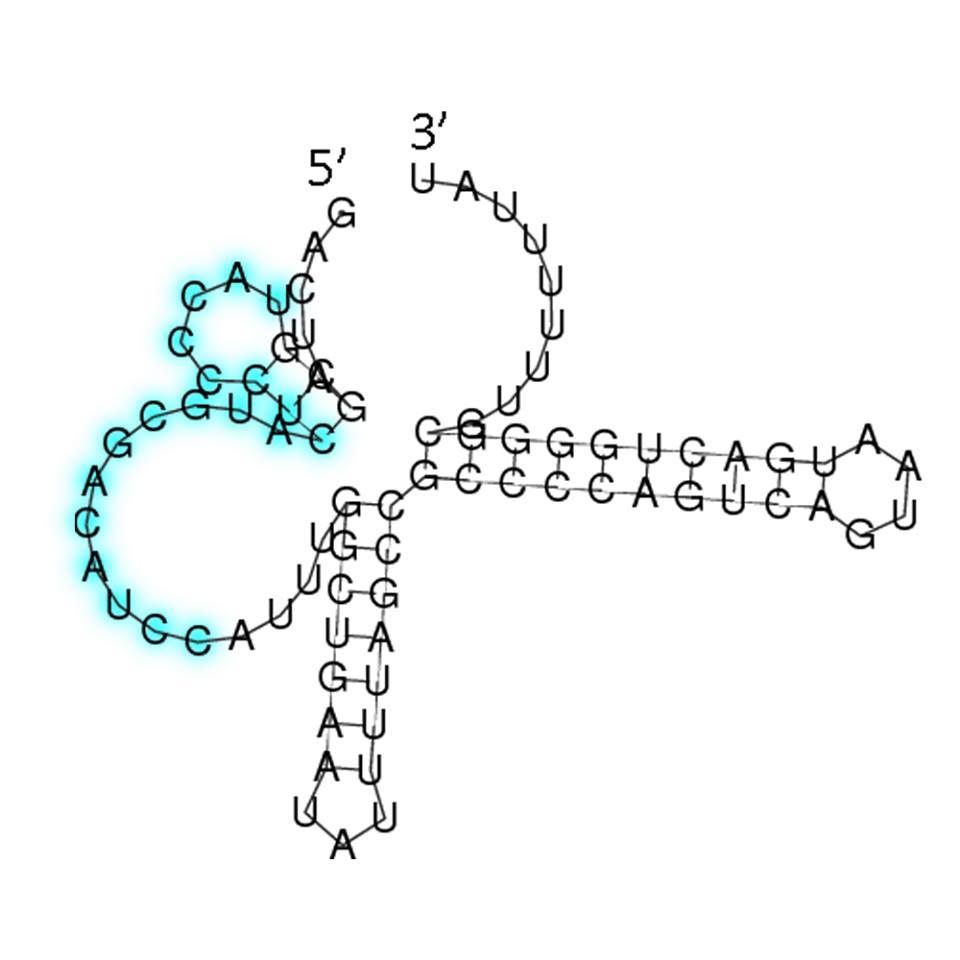
The sRNA picked is competent, but we hoped that it can target any desired gene. Therefore, we designed a RBS by employing the sRNA targeting region from ''OmpF'' and making the AUG codon sufficiently apart from the SD sequence for ribosome binding. By adding this RBS to the upstream of any desired gene, the gene can be regulated by sRNA. | The sRNA picked is competent, but we hoped that it can target any desired gene. Therefore, we designed a RBS by employing the sRNA targeting region from ''OmpF'' and making the AUG codon sufficiently apart from the SD sequence for ribosome binding. By adding this RBS to the upstream of any desired gene, the gene can be regulated by sRNA. | ||
| - | [[File:NCTU_sRNA2.jpg|center|400 px| | + | [[File:NCTU_sRNA2.jpg|center|400 px|Figure 13. The secondary structure of the sRNA-2 we designed]] |
| Line 186: | Line 184: | ||
<li>E.K. Jocelyn, S.G. Elliott , T.K. Stephen, "Lewin's Genes X.-10th ed.", Jones & Bartlett, Sudbury, MA, 2011.</li> | <li>E.K. Jocelyn, S.G. Elliott , T.K. Stephen, "Lewin's Genes X.-10th ed.", Jones & Bartlett, Sudbury, MA, 2011.</li> | ||
<li>Karen M. Wassarman.(2002). "Small RNAs in Bacteria: Diverse Regulators of Gene Expression in Response to Environmental Changes". Cell, 109:141–144</li> | <li>Karen M. Wassarman.(2002). "Small RNAs in Bacteria: Diverse Regulators of Gene Expression in Response to Environmental Changes". Cell, 109:141–144</li> | ||
| - | <li>Hongmarn Park, Geunu Bak, Sun Chang Kim & Younghoon Lee.(2013). "Exploring sRNA-mediated gene silencing mechanisms using artificial small RNAs derived from a natural RNA scaffold in Escherichia coli". Nucleic Acids Research,Vol. 41, No. 6, 3787-3804 DOI:10.1093</li> | + | <li>Hongmarn Park, Geunu Bak, Sun Chang Kim & Younghoon Lee.(2013). "Exploring sRNA-mediated gene silencing mechanisms using artificial small RNAs derived from a natural RNA scaffold in ''Escherichia coli'' ". Nucleic Acids Research,Vol. 41, No. 6, 3787-3804 DOI:10.1093</li> |
<li>Vandana Sharma, Asami Yamamura & Yohei Yokobayashi.(2011). "Engineering Artificial Small RNAs for Conditional Gene Silencing in E. coli". ACS Synthetic Biology</li> | <li>Vandana Sharma, Asami Yamamura & Yohei Yokobayashi.(2011). "Engineering Artificial Small RNAs for Conditional Gene Silencing in E. coli". ACS Synthetic Biology</li> | ||
</ol> | </ol> | ||
| Line 198: | Line 196: | ||
====Introduction==== | ====Introduction==== | ||
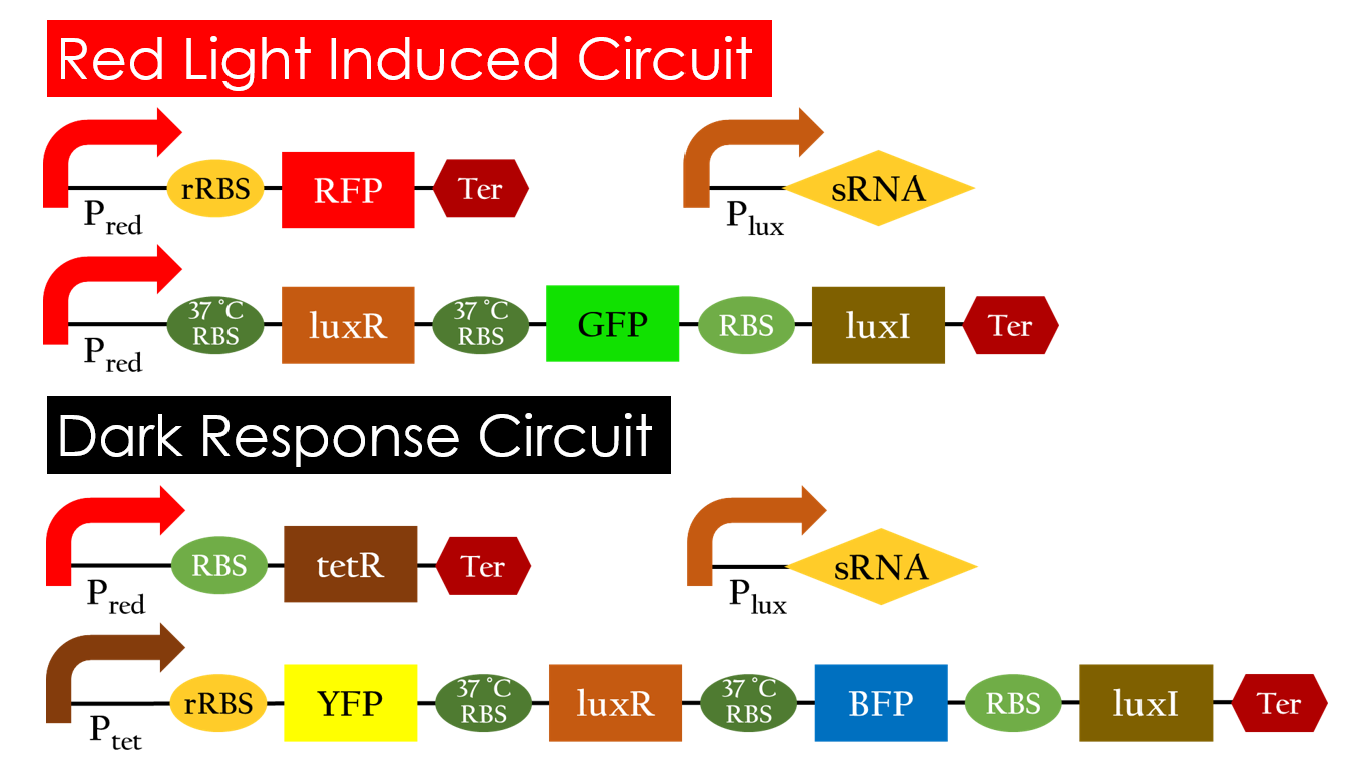
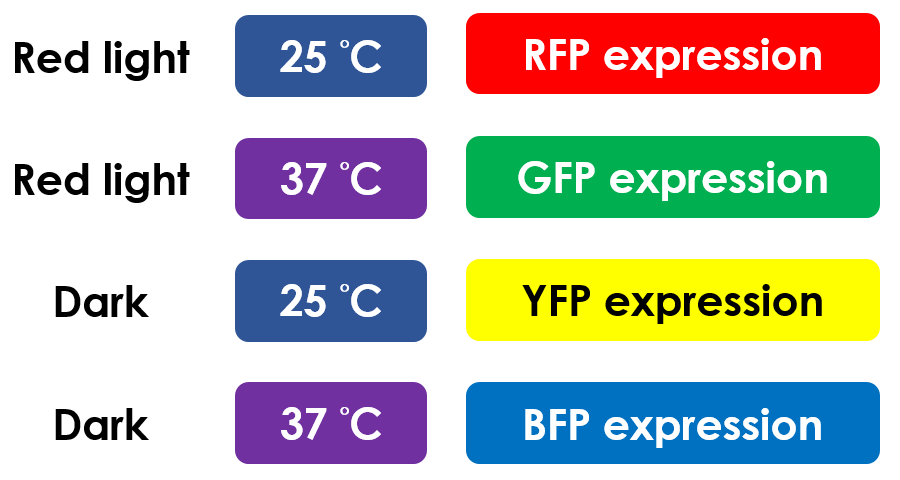
This is the pathway we designed by using the three different regulating factors. We tend to set up four conditions for the ''E.coli'' to work by paring the light and the temperature controls. Distinguished by whether red light exists or not, the pathway is divided into two parts, the '''Red Light Induced Circuit''' and the '''Dark Response Circuit'''. We use four different fluorescent proteins as reporter genes to test that whether the switch of our pathway is successful. | This is the pathway we designed by using the three different regulating factors. We tend to set up four conditions for the ''E.coli'' to work by paring the light and the temperature controls. Distinguished by whether red light exists or not, the pathway is divided into two parts, the '''Red Light Induced Circuit''' and the '''Dark Response Circuit'''. We use four different fluorescent proteins as reporter genes to test that whether the switch of our pathway is successful. | ||
| - | [[File:NCTU_Pathway_Regulation.jpg|center|800px| | + | [[File:NCTU_Pathway_Regulation.jpg|center|800px|Figure 14. The pathway of E.colightuner.]] |
====Red Light Induced Circuit==== | ====Red Light Induced Circuit==== | ||
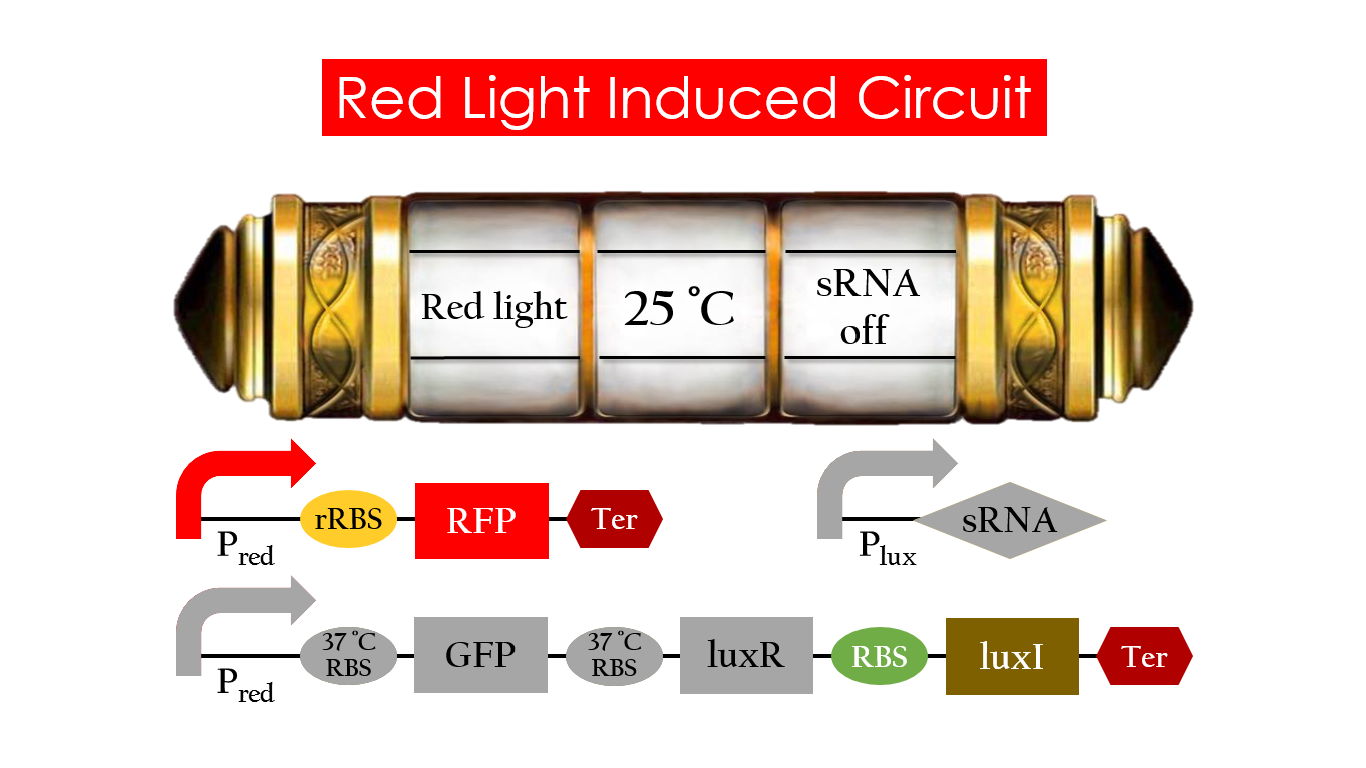
| - | At | + | At 25 °C and under red light, P<sub>red</sub> is activated for translation proceeding. However, ribosomes can only bind to the normal RBS, as 37 °C RBS forms a hairpin structure, forestalling ribosomes from binding. The only gene downstream of the normal RBS is RFP, so only RFP would be expressed. |
| - | [[File:Red_30_off.jpg|center|800px| | + | [[File:Red_30_off.jpg|center|800px|Figure 15. RFP expression under the condition of red light and 25°C.]] |
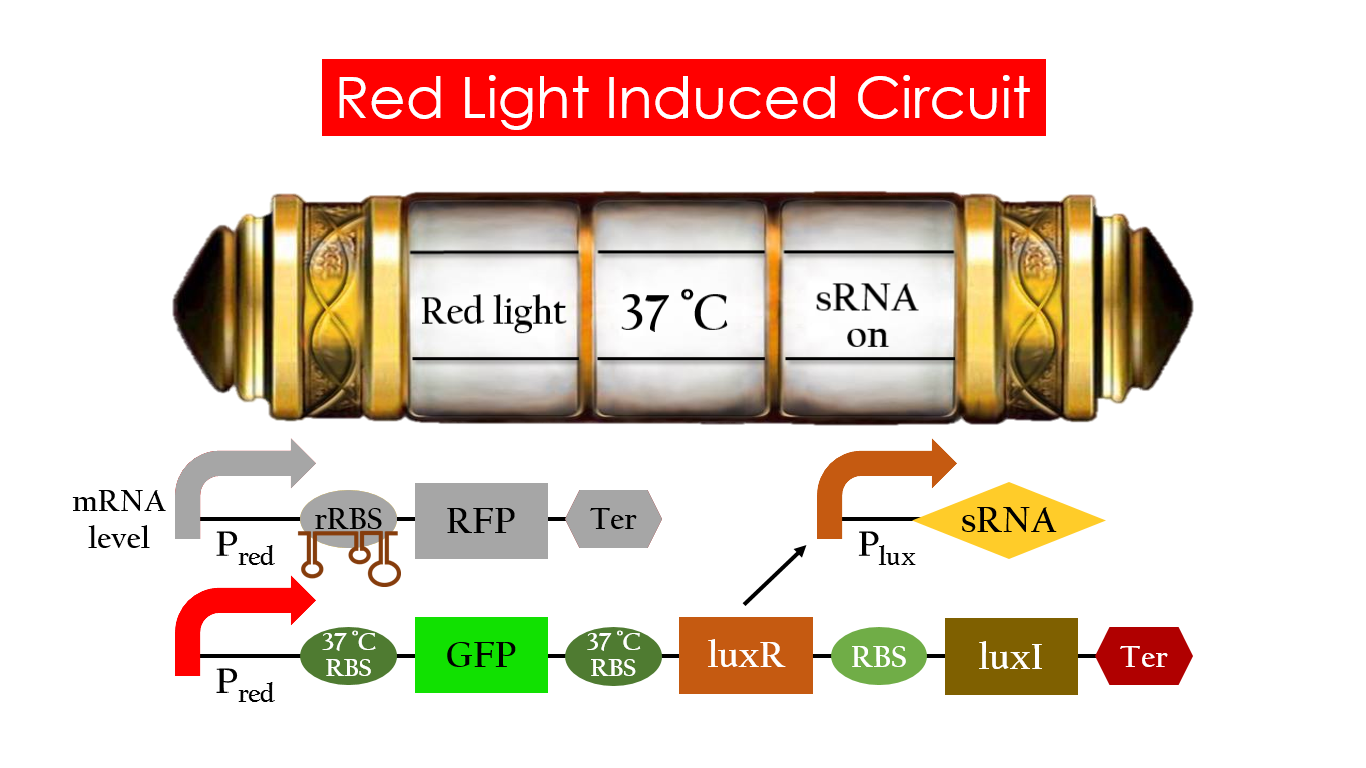
| - | On the other hand, with red light, 37°C RBS unfolds at 37°C , resulting in the expression of both | + | On the other hand, with red light, 37 °C RBS unfolds at 37 °C , resulting in the expression of both LuxR and GFP. LuxR would bind with AHL to form a complex that activates P<sub>lux</sub>. The activation of P<sub>lux</sub> produces sRNA that binds to the normal RBS on mRNA level, blocking it from ribosomes. As a result, RFP would not be expressed and only GFP remains at 37 °C. |
| - | [[File:Red_37_on.jpg|center|800px| | + | [[File:Red_37_on.jpg|center|800px|Figure 16. GFP expression under the condition of red light and 37 °C.]] |
Without red light, P<sub>red</sub> would not be activated, and therefore, the red light induced circuit is completely shut down in the dark. | Without red light, P<sub>red</sub> would not be activated, and therefore, the red light induced circuit is completely shut down in the dark. | ||
====Dark Response Circuit==== | ====Dark Response Circuit==== | ||
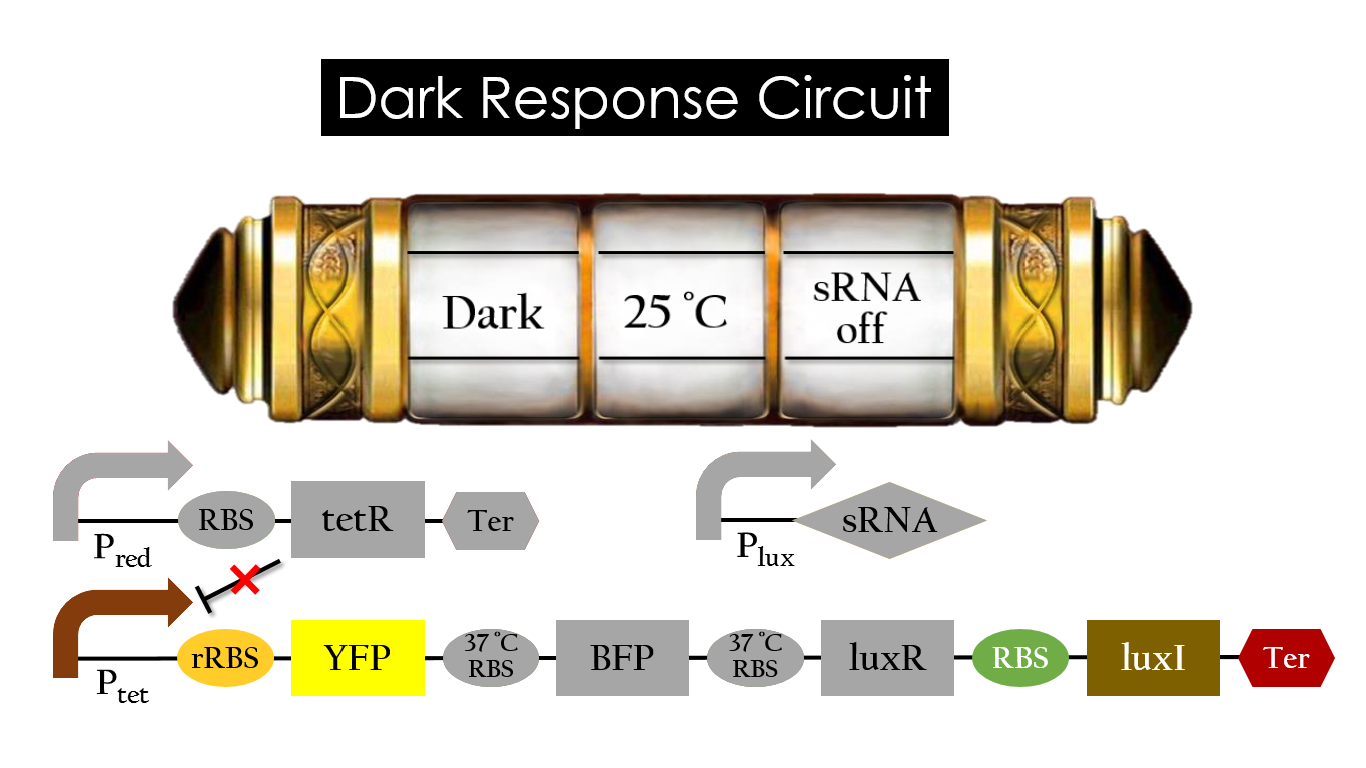
| - | P<sub>red</sub> activates in the presence of red light, producing tetR that represses P<sub>tet</sub>. In other words, the dark induced part is inactive in the exposure to red light and active without red light. At | + | P<sub>red</sub> activates in the presence of red light, producing tetR that represses P<sub>tet</sub>. In other words, the dark induced part is inactive in the exposure to red light and active without red light. At 25 °C in the dark, only normal RBS functions to express YFP while 37 °C RBS remains as a hairpin structure. |
| - | [[File:Dark_30_off.jpg|center|800px| | + | [[File:Dark_30_off.jpg|center|800px|Figure 17. YFP expression under the condition of dark and 25 °C.]] |
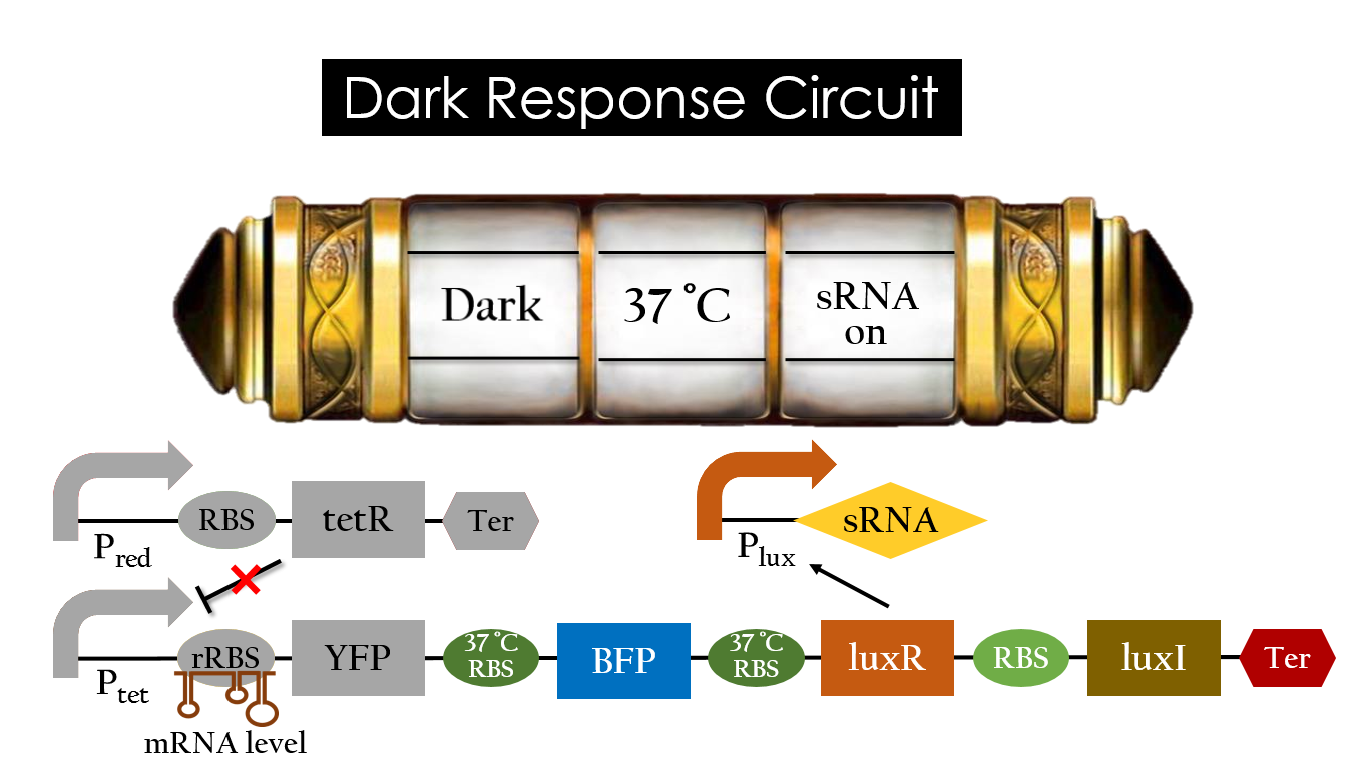
| - | At 37°C, however, 37°C RBS unfolds to express | + | At 37 °C, however, 37 °C RBS unfolds to express LuxR and BFP. Just like the mechanism employed in the light induced part, LuxR forms a complex with AHL to activate P<sub>lux</sub> that produces the sRNA to block normal RBS. This way, the YFP downstream of rRBS cannot be expressed and BFP is expressed at 37 °C. |
| - | [[File:Dark_37_on.jpg|center|800px| | + | [[File:Dark_37_on.jpg|center|800px|Figure 18. BFP expression under the condition of dark and 37 °C.]] |
====Summary==== | ====Summary==== | ||
This is the pathway summary of the E.colightuner. There is one kind of fluorescent protein in each condition we set. It means that we can control four gene expressions by using the E.colightuner. The only thing we have to do is changing the different environmental conditions for specifically expression. | This is the pathway summary of the E.colightuner. There is one kind of fluorescent protein in each condition we set. It means that we can control four gene expressions by using the E.colightuner. The only thing we have to do is changing the different environmental conditions for specifically expression. | ||
| - | [[File:NCTU_Pathway_Regulation_result.png|center||500px| | + | [[File:NCTU_Pathway_Regulation_result.png|center||500px|Figure 19. Summary of E.colightuner.]] |
</div></div> | </div></div> | ||
| + | <div class="li"><div class="card"> | ||
| + | |||
| + | ===i-Culture=== | ||
| + | After a year of hard working, we have manage to accomplish our project modeling, assembling our E.colightuner as well as tissue culturing. The most significant event is we've also combined these three dependent systems and created a magnificent tool, called i-Culture, to achieve the goal of synchronizing different plant growth stages for exportation. We can control our E.colightuner by giving input data into the computer programs, regulating it to secrete the corresponding hormones under specific sets of conditions. | ||
| + | |||
| + | For Example, Taiwan is one of the main exporters of orchid. From a business point of view, the orchid need to blossom precisely at the moment the customers receive it, so the flower growers need to control the blossoms manually during transporting. By using our i-Culture device, we can adjust the blossoming time automatically to reduce the laboring cost. In conclusion, our system has a great development potential in business. | ||
| + | |||
| + | |||
| + | [[File:NCTU_hole_project.png|center|800px|Figure 20. i-Culture, the automation E.colightuner equitment ]] | ||
| + | |||
| + | |||
| + | </div></div> | ||
| + | |||
<div class="li"><div class="card"> | <div class="li"><div class="card"> | ||
===Future Works=== | ===Future Works=== | ||
| - | The multiple regulated-system we have created in this project consists of two different parts: the light induced part and the dark induced part. Each part regulates two different genes, resulting in a total of four genes that can be regulated. We tend to regulate more genes by employing more light sensing promoters. By adding each light sensor, we would be able to regulate two more different genes by employing regulation mechanism of 37°C RBS and sRNA. | + | The multiple regulated-system we have created in this project consists of two different parts : the light induced part and the dark induced part. Each part regulates two different genes, resulting in a total of four genes that can be regulated. We tend to regulate more genes by employing more light sensing promoters. By adding each light sensor, we would be able to regulate two more different genes by employing regulation mechanism of 37 °C RBS and sRNA. |
====Application==== | ====Application==== | ||
| Line 230: | Line 241: | ||
'''Tissue culture''' is the growth of tissues or cells separated from the organism. This is typically facilitated via use of a liquid, semi-solid or solid growth medium, such as broth or agar. Moreover, '''hormones''' are added to the growth medium purposed to what plants need during growth period. However, the hormones needed during each growing state are not the same. So, we need to figure out a way to generate multiple hormones. By adding our engineered ''E.coli'' to the growth medium, the plants can gain multiple hormones secreted by ''E. coli'' under different circumstances. We chose four common hormones as our genes, Auxin, Cytokinin, Gibberellin, and Florigen. | '''Tissue culture''' is the growth of tissues or cells separated from the organism. This is typically facilitated via use of a liquid, semi-solid or solid growth medium, such as broth or agar. Moreover, '''hormones''' are added to the growth medium purposed to what plants need during growth period. However, the hormones needed during each growing state are not the same. So, we need to figure out a way to generate multiple hormones. By adding our engineered ''E.coli'' to the growth medium, the plants can gain multiple hormones secreted by ''E. coli'' under different circumstances. We chose four common hormones as our genes, Auxin, Cytokinin, Gibberellin, and Florigen. | ||
| - | [[File:Nctu_application.png|center|600px| | + | [[File:Nctu_application.png|center|600px|Figure 21. Four hormones and their functions]] |
| - | Since '''Florigen'''(known as the''FT'' protein) is the protein which isn't produced by ''E.coli'' naturally, there aren't any secretion systems in wild type ''E. coli'' to transport Florigen out of the cell. Though secretion through cell lysis may seem as a possible method to solve this problem, we still seek for other ways in order not to let our ''E. coli'' die during the secretion process. Thus, we ought to create a secretion system which can help Florigen pass through the inner and outer membrane of ''E. coli''. We have found a system from the 2012 Kyoto iGEM team called Twin Arginate Translocation (Tat) pathway, which can help send Florigen from the inner membrane to periplasm. In the Tat pathway, a Tat transporter recognizes ''TorA'' signal. Proteins possessing a ''TorA'' signal at their N terminals can permeate into the periplasm. Moreover, the 2012 Kyoto iGEM team also found out that the ''kil'' protein from λ phage are capable of making holes on the outer membrane, which then transports Florigen out of the cell. In other words, with the ''kil'' protein gene fused with the ''torA'' and ''FT'' protein gene in our biobrick, we should be able to successfully transport Florigen out of the ''E. coli''. | + | Since '''Florigen''' (known as the ''FT'' protein) is the protein which isn't produced by ''E.coli'' naturally, there aren't any secretion systems in wild type ''E. coli'' to transport Florigen out of the cell. Though secretion through cell lysis may seem as a possible method to solve this problem, we still seek for other ways in order not to let our ''E. coli'' die during the secretion process. Thus, we ought to create a secretion system which can help Florigen pass through the inner and outer membrane of ''E. coli''. We have found a system from the 2012 Kyoto iGEM team called Twin Arginate Translocation (Tat) pathway, which can help send Florigen from the inner membrane to periplasm. In the Tat pathway, a Tat transporter recognizes ''TorA'' signal. Proteins possessing a ''TorA'' signal at their N terminals can permeate into the periplasm. Moreover, the 2012 Kyoto iGEM team also found out that the ''kil'' protein from λ phage are capable of making holes on the outer membrane, which then transports Florigen out of the cell. In other words, with the ''kil'' protein gene fused with the ''torA'' and ''FT'' protein gene in our biobrick, we should be able to successfully transport Florigen out of the ''E. coli''. |
| - | The use of '''Auxin''' is to induce the growth of shoot, but an excess amount of Auxin would likely kill the plant. We want to reach the maximum amount that will promote the plant’s growth but without killing the plant. After researching, the Imperial College London 2011 iGEM team also had the same problem. They chose the native Auxin indole-3-acetic acid(IAA) as their solution. Compared to the former, the synthetic Auxin is more stable since it can be preserved for weeks. However, an excess amount of IAA could be toxic. To ensure the efficiency but at the same time not excessive, they built a model to reach the maximum amount. | + | The use of '''Auxin''' is to induce the growth of shoot, but an excess amount of Auxin would likely kill the plant. We want to reach the maximum amount that will promote the plant’s growth but without killing the plant. After researching, the Imperial College London 2011 iGEM team also had the same problem. They chose the native Auxin indole-3-acetic acid (IAA) as their solution. Compared to the former, the synthetic Auxin is more stable since it can be preserved for weeks. However, an excess amount of IAA could be toxic. To ensure the efficiency but at the same time not excessive, they built a model to reach the maximum amount. |
We intend to carry out our application with the device beneath. Cultured in the growth medium circulation, our engineered ''E. coli'' can absorb enough nutrition to express the desired genes properly. We also use a '''filter''', which allows hormones but not bacteria themselves to pass through, separating the bacteria from plant tissues. Thus, we keep the tissue culture medium isolated. | We intend to carry out our application with the device beneath. Cultured in the growth medium circulation, our engineered ''E. coli'' can absorb enough nutrition to express the desired genes properly. We also use a '''filter''', which allows hormones but not bacteria themselves to pass through, separating the bacteria from plant tissues. Thus, we keep the tissue culture medium isolated. | ||
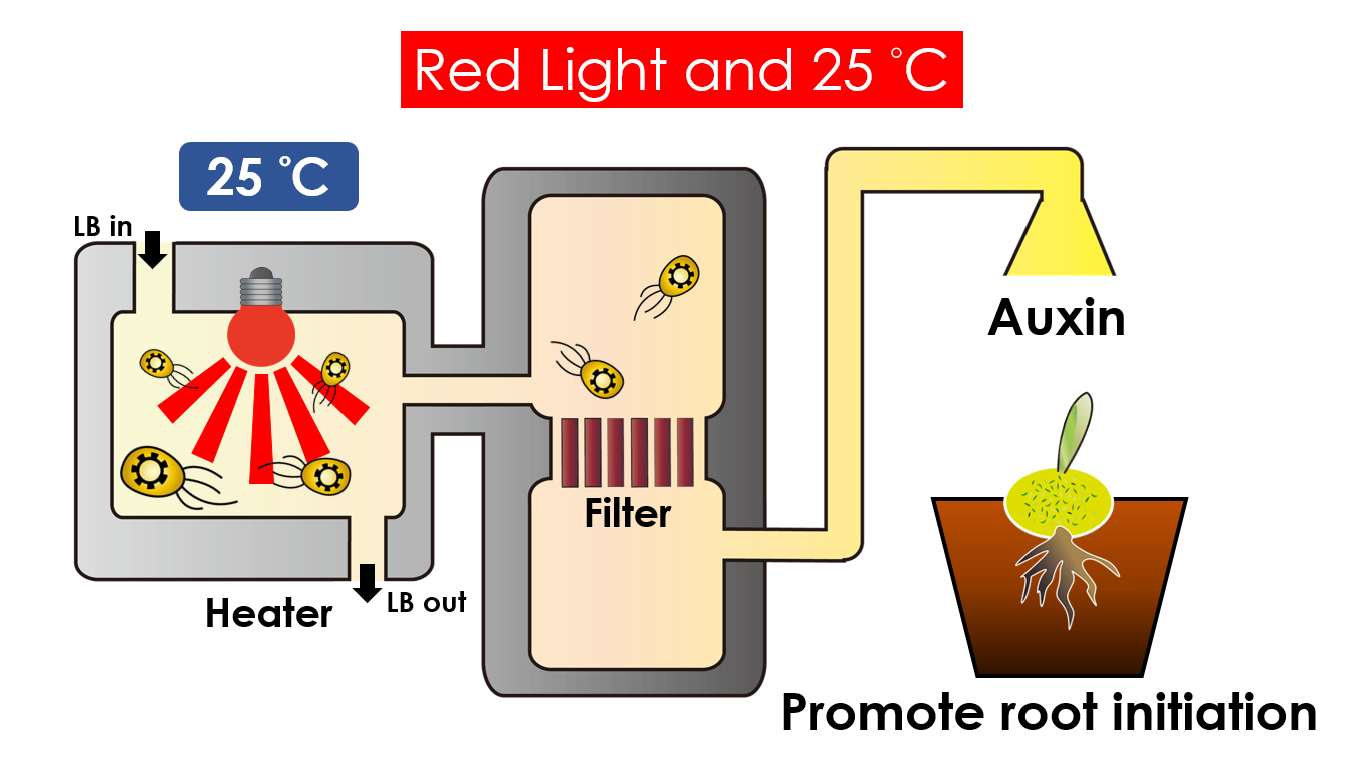
| - | Under red light and | + | Under red light and 37 °C, '''Cytokinin''' will be produced. |
Cytokinin is a plant growth substance which can primarily induce cell growth and differentiation in plant roots and shoots. It can decrease apical dominance which will make the main stem grow better compared with other stems. In this way, all the stems are able to become stronger. Cytokinin can also signal lateral bud growth, which will make the plant bushier. | Cytokinin is a plant growth substance which can primarily induce cell growth and differentiation in plant roots and shoots. It can decrease apical dominance which will make the main stem grow better compared with other stems. In this way, all the stems are able to become stronger. Cytokinin can also signal lateral bud growth, which will make the plant bushier. | ||
| - | [[File:NCTU_application_light_37.png|600px|center| | + | [[File:NCTU_application_light_37.png|600px|center|Figure 22. Cytokinin production under the condition of red light and 37 °C ]] |
| - | Under red light and | + | Under red light and 25 °C, Auxin will be produced. |
Auxin can coordinate development from cellular, organs to the whole plant. Auxin molecules in cells may cause direct responses by stimulating or inhibiting certain gene expressions. In the cellular level, Auxin is important for cell growth. It induces axial elongation in roots and also lateral expansion which can make roots swell. In the whole plant level, it affects the shape of the plants. Because of the different functions, each organ has uneven Auxin concentration and causes specific shapes. | Auxin can coordinate development from cellular, organs to the whole plant. Auxin molecules in cells may cause direct responses by stimulating or inhibiting certain gene expressions. In the cellular level, Auxin is important for cell growth. It induces axial elongation in roots and also lateral expansion which can make roots swell. In the whole plant level, it affects the shape of the plants. Because of the different functions, each organ has uneven Auxin concentration and causes specific shapes. | ||
| - | [[File:NCTU_application_light_30.png|600px|center| | + | [[File:NCTU_application_light_30.png|600px|center|Figure 23. Auxin production under the condition of red light and 25 °C]] |
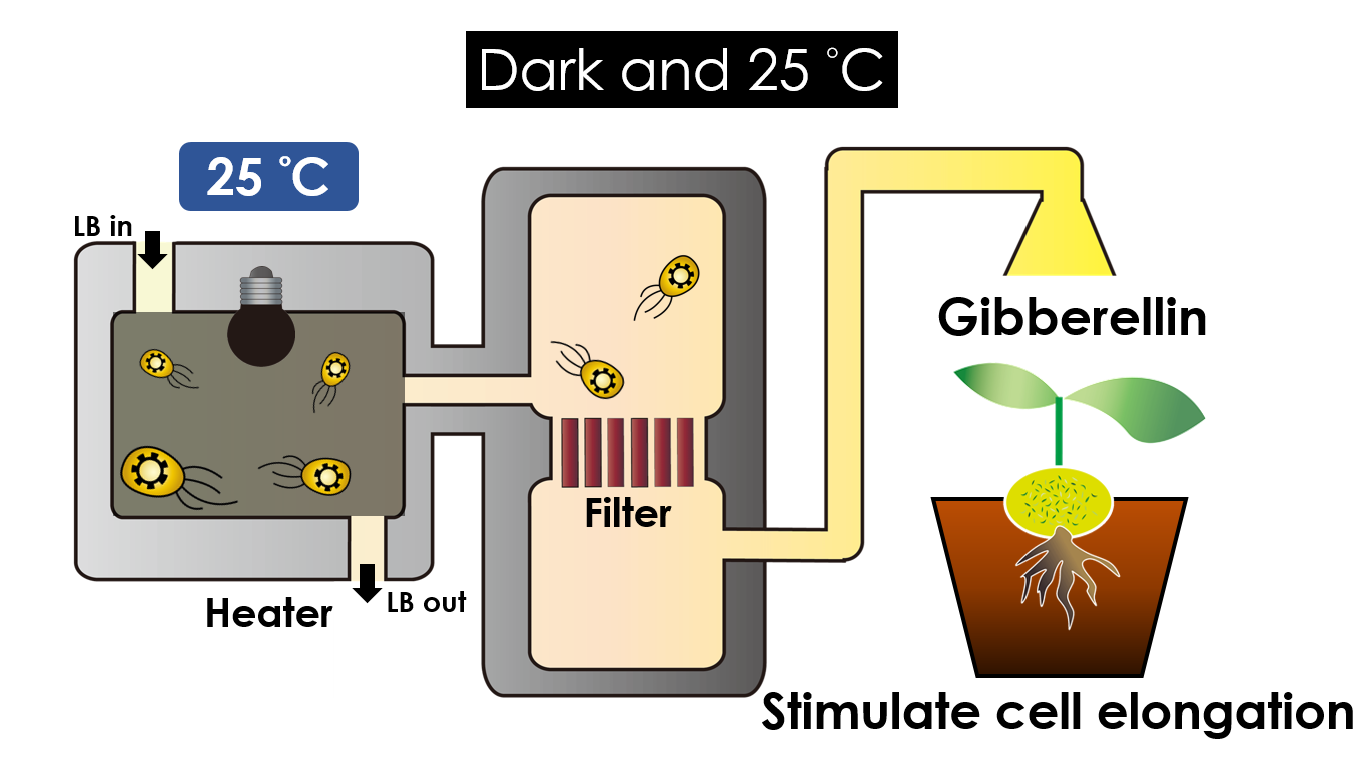
| - | Under dark and | + | Under dark and 25 °C, '''Gibberellin''' will be produced. |
Gibberellin is important in the elongation of cells. It causes the extension of stems by inducing cell divisions and elongations. A special characteristic of it is that it can produce more mass when the plant is exposed to cold temperature. | Gibberellin is important in the elongation of cells. It causes the extension of stems by inducing cell divisions and elongations. A special characteristic of it is that it can produce more mass when the plant is exposed to cold temperature. | ||
| - | [[File:NCTU_application_dark_30.png|600px|center| | + | [[File:NCTU_application_dark_30.png|600px|center|Figure 24. Gibberellin production under the condition of dark and 25 °C]] |
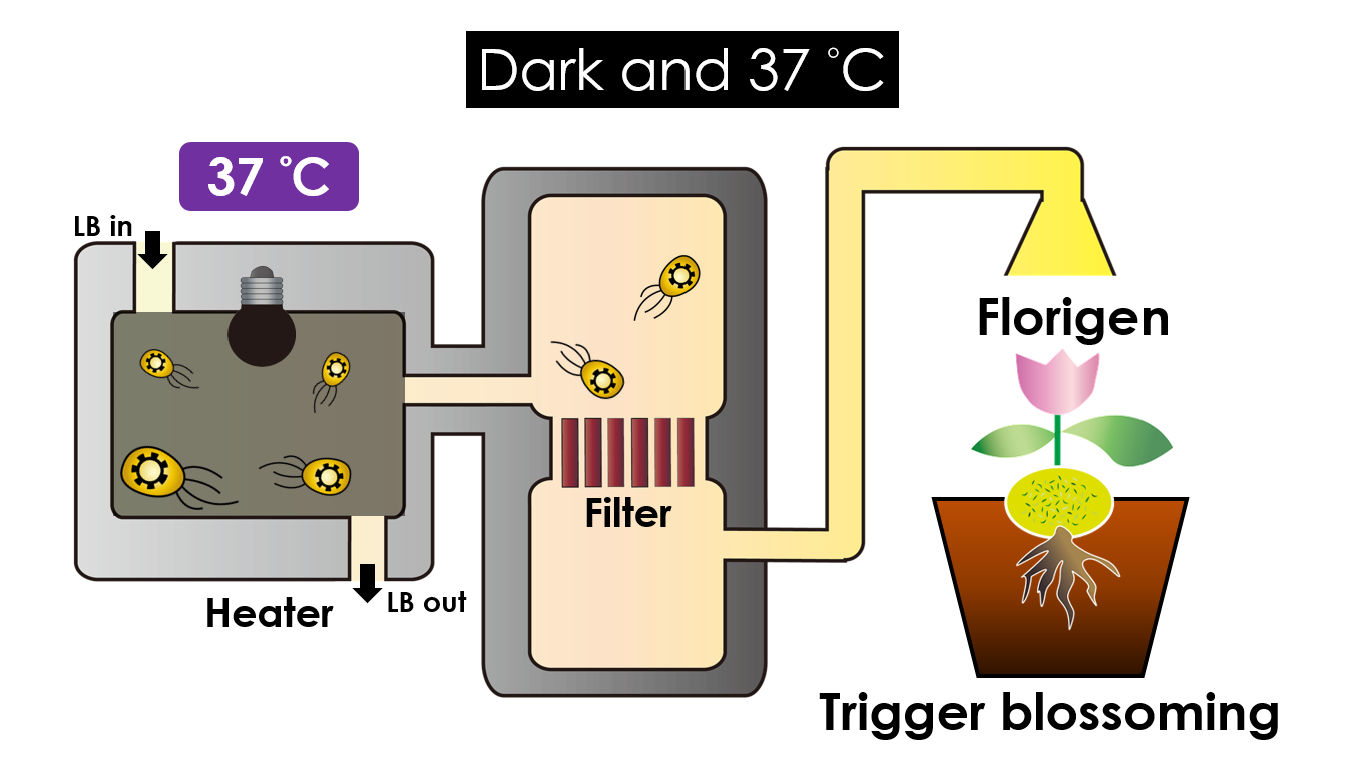
| - | Under dark and | + | Under dark and 37 °C, Florigen will be produced. |
| - | Florigen is the key hormone to trigger plant blossoming. We use a promoter that can promote transcription of the FT gene, which then translate into FT proteins. FT proteins will then be transported via the phloem to the shoot apical meristem, where it will interact with the FD protein(transcription factor) to activate floral identity, thus inducing flowering. | + | Florigen is the key hormone to trigger plant blossoming. We use a promoter that can promote transcription of the FT gene, which then translate into FT proteins. FT proteins will then be transported via the phloem to the shoot apical meristem, where it will interact with the FD protein (transcription factor) to activate floral identity, thus inducing flowering. |
| - | [[File:NCTU_application_dark_37.png|600px|center| | + | [[File:NCTU_application_dark_37.png|600px|center|Figure 25. Florigen production under the condition of dark and 37 °C]] |
====The next BIG thing==== | ====The next BIG thing==== | ||
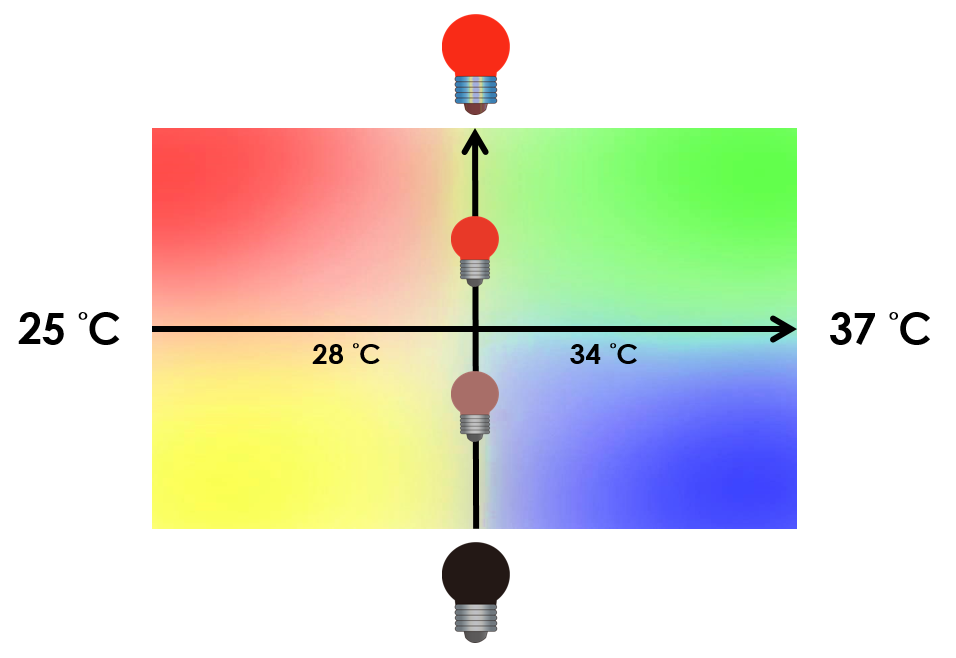
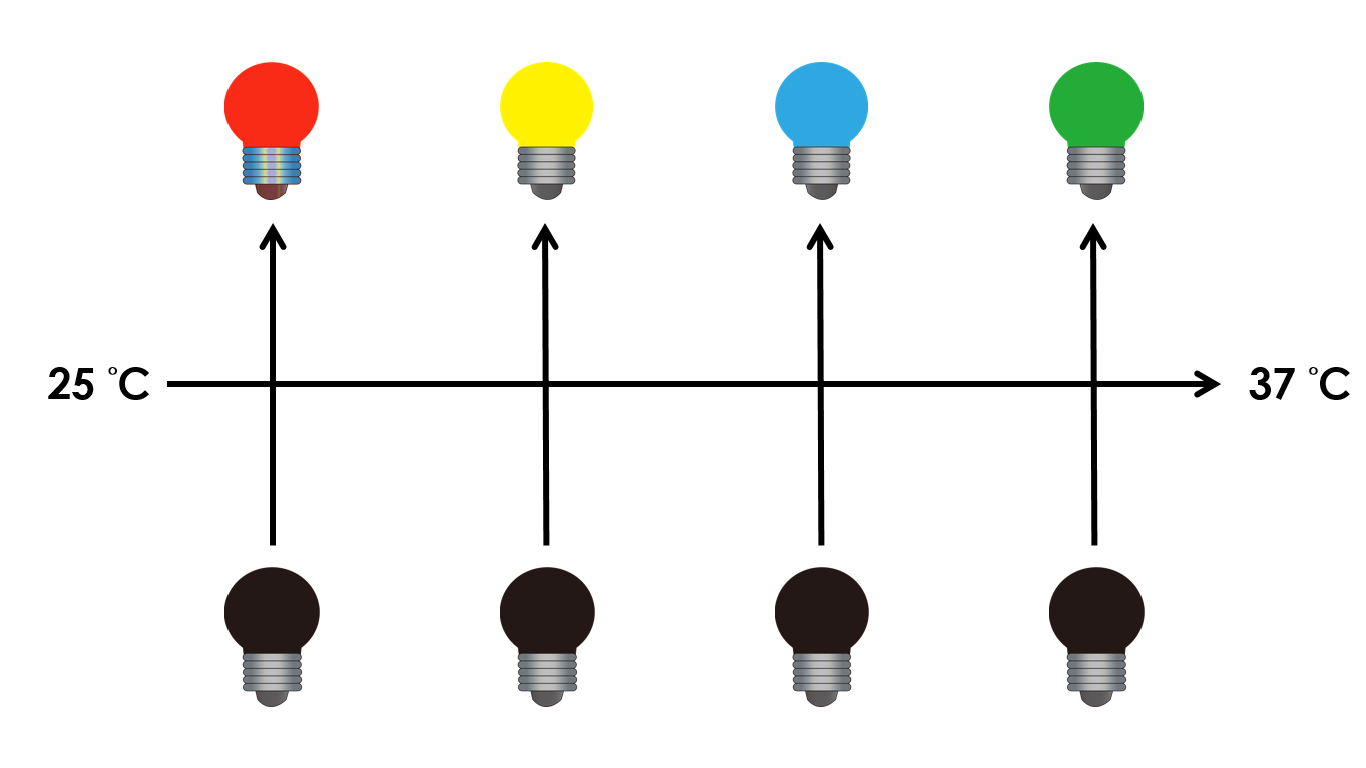
| - | On the other hand, we tend to optimize our system to achieve subtle control of the four gene expressions. We might be able to do this by '''taking the values between the limits we have set'''. Instead of | + | On the other hand, we tend to optimize our system to achieve subtle control of the four gene expressions. We might be able to do this by '''taking the values between the limits we have set'''. Instead of 25 °C and 37 °C , we tend to take the values between, so we can create more conditions under which different level of expressions can be achieved. We can also vary the intensity of red light to control the level of expression. For instance, we can gain different pigment outputs by tuning the temperature and varying the intensity of red light. The ultimate goal is to precisely control the level expression of each gene. |
| - | [[File:NCTU Formosa future work 1.png|800px|center|600px| | + | [[File:NCTU Formosa future work 1.png|800px|center|600px|Figure 26. Different pigment outputs by tuning the temperature between 25 °C and 37 °C and the intensity of red ligtht.]] |
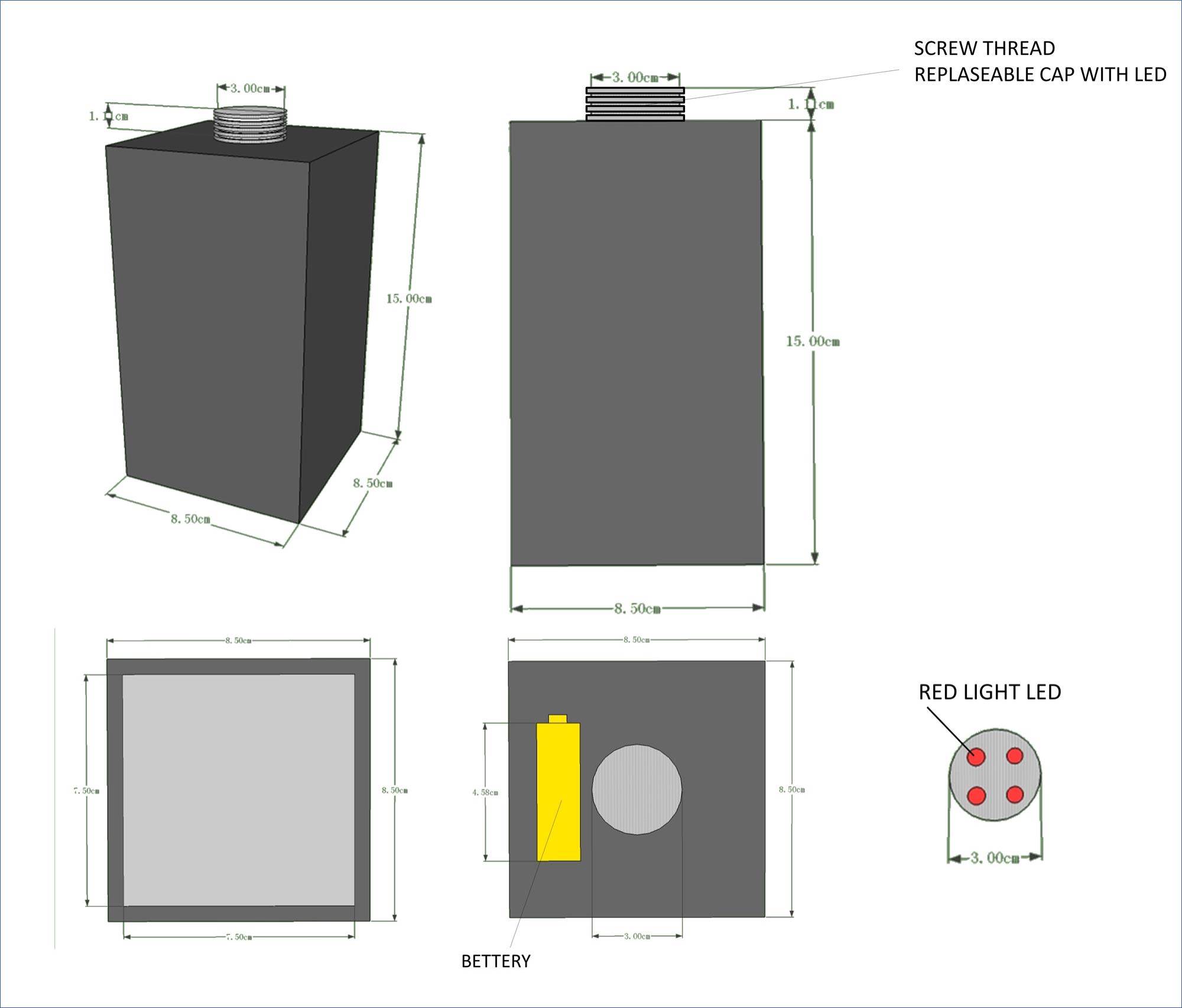
| + | So, we designed and built a device which is able to control the intensity of light. This device is used in the incubator and created for the purpose of covering all the light source outside. So the only light source is the red light LED we put in. In our first design, the device has a cuboid body and a thread top. This thread top is capable to change the light intensity, simply by changing different top with different number of LEDs. | ||
| + | [[File:NCTU FORMOSA WEB WIKI DEVICE DESIGN.JPG|400px|center|Figure 27. The design of the red light control device]] | ||
| + | We use corrugated paper to make a prototype of our light control device. It has one red light LED and the device is covered by several layers of black plastic bag, preventing any possibility of light leaking. | ||
| + | [[File:NCTU FORMOSA WEB WIKI DEVICE1.JPG|400px|center|Figure 28. The prototype of the red light control device]] | ||
As mentioned above in the beginning, using red light as an input condition is an default environment in our project. Our ultimate goal is to regulate multiple genes as more as possible. So, in the future, by employing more different conditions, such as '''different lights''', we would be able to regulate more genes.(one light sensor would result in controlling two more different genes) | As mentioned above in the beginning, using red light as an input condition is an default environment in our project. Our ultimate goal is to regulate multiple genes as more as possible. So, in the future, by employing more different conditions, such as '''different lights''', we would be able to regulate more genes.(one light sensor would result in controlling two more different genes) | ||
We hope that our project could eventually serve as a standardized model and set up with a standard operating procedure which offers future iGEMers to manipulate the control of multiple genes by simply changing the environmental conditions. | We hope that our project could eventually serve as a standardized model and set up with a standard operating procedure which offers future iGEMers to manipulate the control of multiple genes by simply changing the environmental conditions. | ||
| - | [[File:NCTU_app2.png|800px|center|600px| | + | [[File:NCTU_app2.png|800px|center|600px|Figure 29. Add different lights to regulate more genes.]] |
======Reference====== | ======Reference====== | ||
Latest revision as of 03:57, 29 October 2013
A multiple regulated-system was built using three different regulation mechanisms including red light, temperature, and sRNA. In other words, it is multitasking genetic engineered machine that can express a variable genes depending on the different command given.
Contents |
Introduction
The majority of the previous iGEM projects focused on expressing certain genes to achieve a specific task such as sensing a certain phenomenon. This year, we took a step forward in exploring the potential of Escherichia coli, thus building a system that is capable of regulating multiple genes. The system, so-called E.colightuner, is built by three regulated-systems including light regulated-system, temperature regulated-system, and sRNA regulated-system. We used noninvasive factors such as red light and temperature to create different conditions under which E. coli can express different genes. We also introduced a new sRNA regulated-system that hasn't been widely applied in iGEM before.
E.colightuner is just like a machine that can process different tasks depending on the commands given. In this case, the gene expression of E.colightuner is the task and its environmental condition is the command. By changing the environmental condition, E.colightuner can express different genes. The advantage of building such a machine is to increase the convenience in expressing multiple genes. Instead of expressing different genes with different bacteria, we can express the genes with E.colightuner. This may ultimately lower the cost in expressing different genes.
We tend to apply E.colightuner to agriculture. By expressing different hormones required by different stages of plant growth, E.colightuner can aid plant growth. With that said, we built a device that can flower the desired hormones to plants. This device can be applied to any plantation, and it is simple to manipulate. With more efforts, we believe that E.colightuner can raise the potential values of E.coli.
Light-regulated system
Introduction
Light is an easily accessible energy that can be applied to various fields. Of course, it can be applied to synthetic biology as well. Bacteria can sense light of a defined wavelength through a small light-absorbing ligand called chromophore. By employing this chromophore, light can be used to trigger a certain reaction and even regulate gene expressions.
In E.colightuner, light serves as an activator that can regulate the function of a promoter. Since red light can be easily turned on and off, the activation and repression of the promoter can also be easily achieved. Substrate activators, on the other hand, are harder to manipulate. Substrates can activate their corresponding promoters efficiently, but they might be hard to remove, making it difficult to repress the promoters when desired.
In order to make E. coli, sense and respond to light, we installed a "two component system" (cph8) that can respond to red light.
The chimera cph8 is consisted of a phytochrome cph1 and a histidine kinase domain, envz-ompR, that includes a response regulator. cph1 is the first member of the plant photoreceptor family that has been identified in bacteria. The functional expression of a phytochrome domain (cph1) in E. coli requires the biosynthesis of the respective bilin chromophore PCB. Two phycocyanobilin(PCB)-biosynthesis genes (ho11 and pcya2) from Synechocystis encoding a heme oxygenase and bilin reductase, respectively, is essential to convert heme into PCB. Figure 2 depicts the phycocyanobilin-biosynthesis pathway.3 With the biosynthesis of PCB, cph1 serve as a red-light absorbing chromophore that is inactivated under red light and activated without red light . 4,5
envZ-ompR, a dimeric osmosensor, is a multidomain transmembrane protein and one of the best characterized two-component histidine kinases from E.coli. Besides the periplasmic receptor domain that is flanked by two transmembrane helices, it possesses a C-terminal 228-residue histidine kinase domain that is located in the cytoplasm6. Upon changes of extracellular osmolarity, envZ specifically phosphorylates its cognate response regulator ompR, which, in turn, regulates the Pompc 7.
With the two components just mentioned, cph8 can serve as a photoreceptor that regulates gene expression through Pompc. Without red light, cph1 is activated and it enables envZ-ompR to autophosphorylate which in turn activates Pompc. Under the exposure of red light, however, cph1 is deactivated, inhibiting the autophosphorylation, thus turning off gene expression. This mechanism is depicted in Figure 38 and the experimental data that support this mechanism is shown in Figure 4.
Design of Red promoter
We designed a new biobrick, Pred, to positively regulate gene expression using red light. Compared to the negative regulation provided by cph8, positive regulation is more direct and convenient to operate. In addition, cph8 relies on Pompc to express downstream gene, but our design relies on a stronger promoter, Plac, that can amplify gene expression, further making sure that the level of expression can meet our needs.
In the presence of red light, Pompc is shut down and lacI cannot be expressed to repress Plac. As a result, Plac is activated and all the downstream genes can be transcribed. In the absence of red light, whatsoever, Pompc is activated as autophosphorylation is allowed. LacI can be expressed to repress Plac and the genes downstream cannot be expressed, consequently. By using Pred, we can achieve positive control using red light.
Reference
- part BBa_I15008;MIT Registry of Standard Biological Parts
- part BBa_I15009;MIT Registry of Standard Biological Parts
- Levskaya, A. et al .(2005). Engineering Escherichia coli to see light. Nature, 438(7067), 442.
- Kehoe DM, Grossman AR (1996) Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273(5280):1409–1412
- Yeh KC, Wu SH, Murphy JT, Lagarias JC (1997) A cyanobacterial phytochrome two-component light sensory system. cience 277 (5331):1505–1508
- Dutta R, Qin L, Inouye M (1999) Histidine kinases: diversity of domain organization. Mol Microbiol 34(4):633–640
- Forst SA, Roberts DL (1994) Signal transduction by the EnvZ–OmpR phosphotransfer system in bacteria. Res Microbiol 45(5–6):363–373
- Thomas Drepper, Ulrich Krauss,Sonja Meyer zu Berstenhorst, Jörg Pietruszka, Karl-Erich Jaeger.(2011).Lights on and action! Controlling microbial gene expression by light. Appl Microbiol Biotechnol, 90:23–40 DOI:10.1007/s00253-011-3141-6
Temperature-regulated system
Introduction
It is not a dream anymore that "making the impossible possible" with the increase of modern technology. It is also a serious problem that what human has brought to Earth along with our progressing civilization. Drastic environmental changes are inevitable due to the fact of massive developing industries. Temperature changing are one of the problems we need to confront.
Cells are constantly subjected to changing environmental conditions. Comparing with chemical substances, temperature is more accessible, convenient and lower costing. Moreover, temperature is also less burden to the organisms. Therefore, few organisms use temperature as a regulating factor. A mechanism found in different organisms that makes the cell respond to thermal changes, for example, is the RNA thermometer.
An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. The expression of heat-shock, cold-shock and some virulence genes are coordinated in response to temperature changes. There are several systems suggested in references that are based on RNA secondary structure. This structural transition can then expose or occlude important regions of RNA such as a ribosome binding site, which then affects the translation rate of a nearby protein-coding gene, so we can say that the RNA thermometers are just like riboswitches.
Apart from protein-mediated transcriptional control mechanisms, translational control by RNA thermometers is also a widely used regulatory strategy.
Mechanism
RNA thermometers are identified in the 5' UTR of messenger RNA, upstream of a protein-coding gene.
Unlike normal RBS, the 37 °C RBS has a unique or rather stable hairpin structure which shaped like a lock. If the temperature drops below 37 °C, the base-pairing neucleotides on the RNA will form a stable secondary hairpin structure, occluding the Shine-Dalgarno sequence (SD sequence) thus disabling the ribosome to bind. The base-pairing of this RNA region will block the expression of the protein encoded within. Since the structure is sustained by base pairing, heat can be employed to break the hydrogen bonds. So when it reaches 37 °C, the bonds would be broken and the hairpin would be unfolded, causing the SD sequence to be exposed and permitting the binding of the ribosome, which then starts the translation. This means that by raising temperature to a certain threshold, the 37 °C RBS can function as a normal RBS. In this way, gene expression can be regulated on the RNA level by temperature.
Design
The specific ribosome binding site (RBS) [http://parts.igem.org/Part:BBa_K115002 BBa_ K115002] is a great RNA thermometer. It has high translation activity at high temperature (> 37 °C) and low translation activity at room temperature. It was designed for the temperature-dependent genetic circuit in E. coli, with a green fluorescent protein (GFP) used as the reporter protein.
Originally, it is in the 5'-UTR of the Salmonella agsA gene, which codes for a small heat shock protein that can be used for temperature sensitive post-transcriptional regulation, initiating translation at 37 °C. Transcription of the agsA gene is controlled by the alternative sigma factor σ32. Additional translational control depends on a stretch of four uridines that pair with the SD sequence, so they are named "FourU". They repress translation of this protein by base-pairing with the Shine-Dalgardo sequence of the mRNA. This prevents ribosomes from binding the start codon.
In our project, we use the 37 °C RBS in both red light induced circuit and dark response circuit and it is related to the production of GFP and BFP. We use this 37 °C RBS to design our project because we want to use different conditions to produce different substances. Therefore, the characteristic and sensitivity of 37 °C RBS perfectly fits our purpose which we want to regulate our system through temperature.
Reference
- Torsten Waldminghaus, Nadja Heidrich, Sabine Brantl and Franz Narberhaus .(2007). FourU: a novel type of RNA thermometer in Salmonella . Molecular Microbiology , 65(2): 413–424 DOI:10.1111/j.1365-2958.2007.05794.x
- part BBa_K115002;TUDelft Registry of Standard Biological Parts
Small RNA-regulated system
Introduction
Base pairing offers a powerful way for one RNA to control the activity of another. Both prokaryotes and eukaryotes have many cases which a single-stranded RNA base pairs with a complementary region of an mRNA. As a result, it prevents expression of the mRNA.
RNA interference (RNAi), also called post transcriptional gene silencing, is a process in which RNA molecules inhibit gene expression by destroying specific mRNA molecules. In 2006, Andrew Fire and Craig C.Mello shared the Nobel Prize in Physiology or Medicine because of their study on RNA interference11. This powerful gene silencing tool in eukaryotes has been used in many research. As their counterparts in bacteria, small non-coding RNAs (sRNAs) are important regulatory roles.
Small RNAs (sRNAs) have become increasingly significant in playing the role of bacterial gene regulation. Most sRNAs interact with the targeted mRNAs by imperfect base pairing, reducing the translation efficiency and recruiting chaperones such as Hfq for translation termination. The sRNAs would bind to the target with its hairpin-like structure which wound by some of it's own sequences, and the chaperons would stuck between the hairpins in order to protect the sRNA-mRNA combination from degrading. In the end, the sRNAs regulate gene expression by forestalling translation. Since this regulated-system has already been employed in vivo, it is necessary to design an artificial sRNA that targets specifically to a desired genes, so that we can prevent the sRNA from affecting other undesired genes.
Unlike the regulated-system popular used in iGEM projects (e.g. Ptet & tetR, Plux & LuxR, etc.), sRNA regulated-system seems more efficient. Because the inhibition works under RNA level, which means the energy wasted in E. coli is less than other system (producing proteins to regulate).
Figure 8 depicts how the small RNA chaperone, Hfq, associates with the small RNA and represses a target mRNA.12
Mechanism
How do small RNAs regulate gene expression? Figure 9 shows the mechanism of repressing one target mRNA. The small RNA has three stem-loop double stranded RNA structures, and the loop which is closest to the 3’ terminus is complementary to the sequence preceding the initiation codon of the target mRNA. Base pairing between small RNA and the target mRNA prevents the ribosome from binding to the initiation codon, so the translation would be repressed.13,14
Design
The following is the process of our idea in designing the sRNA.
We have designed two artificial sRNAs, each modified from different paper. The sRNA-1 is based on afsRNA ARlacZ1, which is created by Hongmarn Park and his colleagues.15 SibC is a small RNA sequence which is already found in E.coli. Hongmarn Park and his colleagues changed few base pairs of SibC to make ARlacZ1, purposed to make the secondary structure more stable. After that, they tried to change 9 different locations of the target-recognition sequence to test the effect on gene silencing. Our design is to modify from the RNA sequence which has the best efficiency : ARlacZ7. In accordance with afsRNA ARlacZ7, we changed the target-recognition sequence to finally complete our sRNA-1.
The other sRNA sequence we produced is called sRNA-2. It is employed in this project selected from a library of artificial sRNA that was constructed by fusing a randomized antisense domain of Spot42, the scaffold that is known for recruiting the RNA chaperons. The sRNA we picked contains a consensus sequence, 5’-CCCUC-3’, which can base pair with the SD sequence due to complementary binding. This sRNA, as expected, effectively regulates gene expression by reducing translation efficiency and recruiting Hfq. In addition, this sRNA shows high specificity against its targeted gene, OmpF, as it doesn't hold significant activity against other genes from E. coli genome (Sharma and others, 2011).
The sRNA picked is competent, but we hoped that it can target any desired gene. Therefore, we designed a RBS by employing the sRNA targeting region from OmpF and making the AUG codon sufficiently apart from the SD sequence for ribosome binding. By adding this RBS to the upstream of any desired gene, the gene can be regulated by sRNA.
Reference
- Xu, S.; Montgomery, M.; Kostas, S.; Driver, S.; Mello, C. (1998). "Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans". Nature 391 (6669): 806–811 DOI:10.1038/35888
- Jörg Vogel , Ben F. Luisi.(2011). Hfq and its constellation of RNA. Nature Reviews Microbiology, 9:578-589
- E.K. Jocelyn, S.G. Elliott , T.K. Stephen, "Lewin's Genes X.-10th ed.", Jones & Bartlett, Sudbury, MA, 2011.
- Karen M. Wassarman.(2002). "Small RNAs in Bacteria: Diverse Regulators of Gene Expression in Response to Environmental Changes". Cell, 109:141–144
- Hongmarn Park, Geunu Bak, Sun Chang Kim & Younghoon Lee.(2013). "Exploring sRNA-mediated gene silencing mechanisms using artificial small RNAs derived from a natural RNA scaffold in Escherichia coli ". Nucleic Acids Research,Vol. 41, No. 6, 3787-3804 DOI:10.1093
- Vandana Sharma, Asami Yamamura & Yohei Yokobayashi.(2011). "Engineering Artificial Small RNAs for Conditional Gene Silencing in E. coli". ACS Synthetic Biology
Mechanism of our project
Introduction
This is the pathway we designed by using the three different regulating factors. We tend to set up four conditions for the E.coli to work by paring the light and the temperature controls. Distinguished by whether red light exists or not, the pathway is divided into two parts, the Red Light Induced Circuit and the Dark Response Circuit. We use four different fluorescent proteins as reporter genes to test that whether the switch of our pathway is successful.
Red Light Induced Circuit
At 25 °C and under red light, Pred is activated for translation proceeding. However, ribosomes can only bind to the normal RBS, as 37 °C RBS forms a hairpin structure, forestalling ribosomes from binding. The only gene downstream of the normal RBS is RFP, so only RFP would be expressed.
On the other hand, with red light, 37 °C RBS unfolds at 37 °C , resulting in the expression of both LuxR and GFP. LuxR would bind with AHL to form a complex that activates Plux. The activation of Plux produces sRNA that binds to the normal RBS on mRNA level, blocking it from ribosomes. As a result, RFP would not be expressed and only GFP remains at 37 °C.
Without red light, Pred would not be activated, and therefore, the red light induced circuit is completely shut down in the dark.
Dark Response Circuit
Pred activates in the presence of red light, producing tetR that represses Ptet. In other words, the dark induced part is inactive in the exposure to red light and active without red light. At 25 °C in the dark, only normal RBS functions to express YFP while 37 °C RBS remains as a hairpin structure.
At 37 °C, however, 37 °C RBS unfolds to express LuxR and BFP. Just like the mechanism employed in the light induced part, LuxR forms a complex with AHL to activate Plux that produces the sRNA to block normal RBS. This way, the YFP downstream of rRBS cannot be expressed and BFP is expressed at 37 °C.
Summary
This is the pathway summary of the E.colightuner. There is one kind of fluorescent protein in each condition we set. It means that we can control four gene expressions by using the E.colightuner. The only thing we have to do is changing the different environmental conditions for specifically expression.
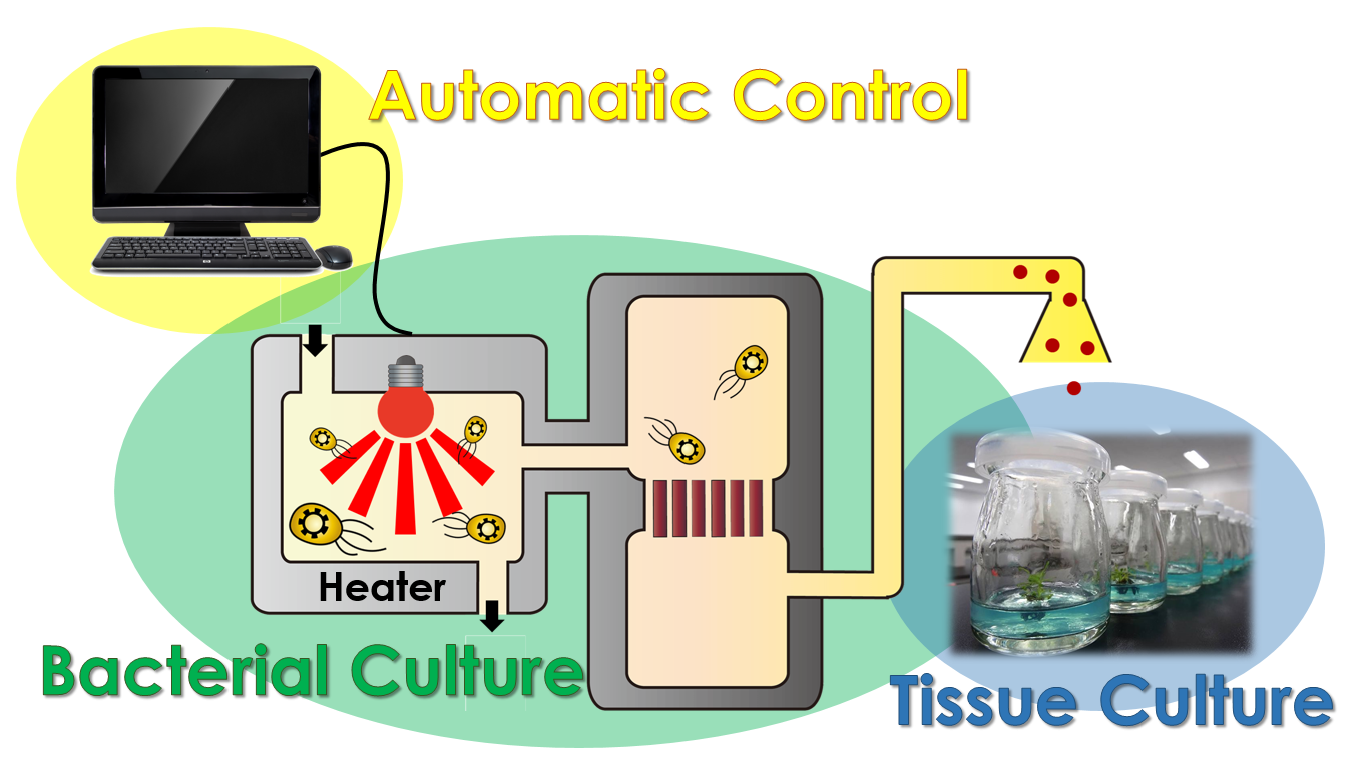
i-Culture
After a year of hard working, we have manage to accomplish our project modeling, assembling our E.colightuner as well as tissue culturing. The most significant event is we've also combined these three dependent systems and created a magnificent tool, called i-Culture, to achieve the goal of synchronizing different plant growth stages for exportation. We can control our E.colightuner by giving input data into the computer programs, regulating it to secrete the corresponding hormones under specific sets of conditions.
For Example, Taiwan is one of the main exporters of orchid. From a business point of view, the orchid need to blossom precisely at the moment the customers receive it, so the flower growers need to control the blossoms manually during transporting. By using our i-Culture device, we can adjust the blossoming time automatically to reduce the laboring cost. In conclusion, our system has a great development potential in business.
Future Works
The multiple regulated-system we have created in this project consists of two different parts : the light induced part and the dark induced part. Each part regulates two different genes, resulting in a total of four genes that can be regulated. We tend to regulate more genes by employing more light sensing promoters. By adding each light sensor, we would be able to regulate two more different genes by employing regulation mechanism of 37 °C RBS and sRNA.
Application
Taiwan is renowned for Phalaenopsis, producing more than 35 million Phalaenopsis each year. There is no doubt that Taiwan has become "The Kingdom of Phalaenopsis". The technique we usually use to grow Phalaenopsis is plant tissue culture, thus we intend to apply it to our system.
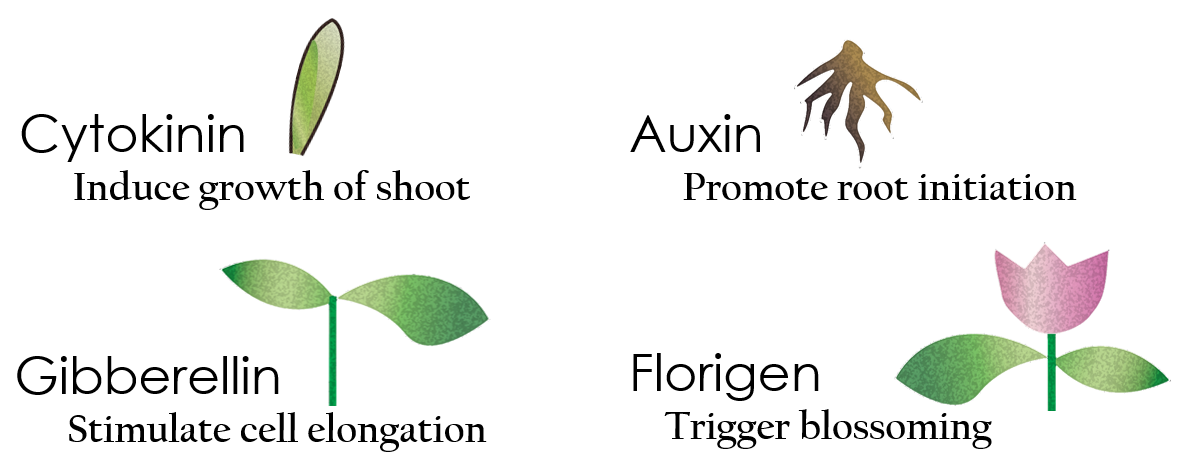
Tissue culture is the growth of tissues or cells separated from the organism. This is typically facilitated via use of a liquid, semi-solid or solid growth medium, such as broth or agar. Moreover, hormones are added to the growth medium purposed to what plants need during growth period. However, the hormones needed during each growing state are not the same. So, we need to figure out a way to generate multiple hormones. By adding our engineered E.coli to the growth medium, the plants can gain multiple hormones secreted by E. coli under different circumstances. We chose four common hormones as our genes, Auxin, Cytokinin, Gibberellin, and Florigen.
Since Florigen (known as the FT protein) is the protein which isn't produced by E.coli naturally, there aren't any secretion systems in wild type E. coli to transport Florigen out of the cell. Though secretion through cell lysis may seem as a possible method to solve this problem, we still seek for other ways in order not to let our E. coli die during the secretion process. Thus, we ought to create a secretion system which can help Florigen pass through the inner and outer membrane of E. coli. We have found a system from the 2012 Kyoto iGEM team called Twin Arginate Translocation (Tat) pathway, which can help send Florigen from the inner membrane to periplasm. In the Tat pathway, a Tat transporter recognizes TorA signal. Proteins possessing a TorA signal at their N terminals can permeate into the periplasm. Moreover, the 2012 Kyoto iGEM team also found out that the kil protein from λ phage are capable of making holes on the outer membrane, which then transports Florigen out of the cell. In other words, with the kil protein gene fused with the torA and FT protein gene in our biobrick, we should be able to successfully transport Florigen out of the E. coli.
The use of Auxin is to induce the growth of shoot, but an excess amount of Auxin would likely kill the plant. We want to reach the maximum amount that will promote the plant’s growth but without killing the plant. After researching, the Imperial College London 2011 iGEM team also had the same problem. They chose the native Auxin indole-3-acetic acid (IAA) as their solution. Compared to the former, the synthetic Auxin is more stable since it can be preserved for weeks. However, an excess amount of IAA could be toxic. To ensure the efficiency but at the same time not excessive, they built a model to reach the maximum amount.
We intend to carry out our application with the device beneath. Cultured in the growth medium circulation, our engineered E. coli can absorb enough nutrition to express the desired genes properly. We also use a filter, which allows hormones but not bacteria themselves to pass through, separating the bacteria from plant tissues. Thus, we keep the tissue culture medium isolated.
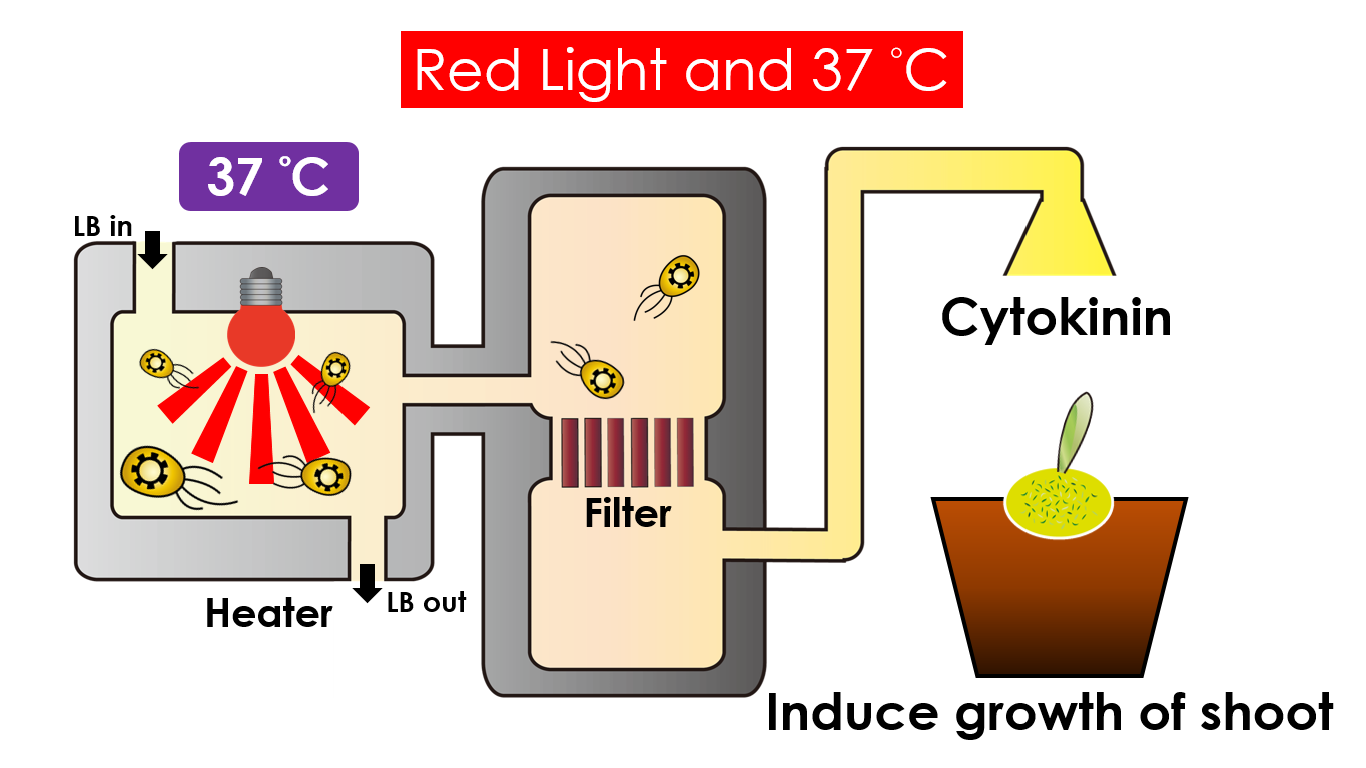
Under red light and 37 °C, Cytokinin will be produced. Cytokinin is a plant growth substance which can primarily induce cell growth and differentiation in plant roots and shoots. It can decrease apical dominance which will make the main stem grow better compared with other stems. In this way, all the stems are able to become stronger. Cytokinin can also signal lateral bud growth, which will make the plant bushier.
Under red light and 25 °C, Auxin will be produced. Auxin can coordinate development from cellular, organs to the whole plant. Auxin molecules in cells may cause direct responses by stimulating or inhibiting certain gene expressions. In the cellular level, Auxin is important for cell growth. It induces axial elongation in roots and also lateral expansion which can make roots swell. In the whole plant level, it affects the shape of the plants. Because of the different functions, each organ has uneven Auxin concentration and causes specific shapes.
Under dark and 25 °C, Gibberellin will be produced. Gibberellin is important in the elongation of cells. It causes the extension of stems by inducing cell divisions and elongations. A special characteristic of it is that it can produce more mass when the plant is exposed to cold temperature.
Under dark and 37 °C, Florigen will be produced. Florigen is the key hormone to trigger plant blossoming. We use a promoter that can promote transcription of the FT gene, which then translate into FT proteins. FT proteins will then be transported via the phloem to the shoot apical meristem, where it will interact with the FD protein (transcription factor) to activate floral identity, thus inducing flowering.
The next BIG thing
On the other hand, we tend to optimize our system to achieve subtle control of the four gene expressions. We might be able to do this by taking the values between the limits we have set. Instead of 25 °C and 37 °C , we tend to take the values between, so we can create more conditions under which different level of expressions can be achieved. We can also vary the intensity of red light to control the level of expression. For instance, we can gain different pigment outputs by tuning the temperature and varying the intensity of red light. The ultimate goal is to precisely control the level expression of each gene.
So, we designed and built a device which is able to control the intensity of light. This device is used in the incubator and created for the purpose of covering all the light source outside. So the only light source is the red light LED we put in. In our first design, the device has a cuboid body and a thread top. This thread top is capable to change the light intensity, simply by changing different top with different number of LEDs.
We use corrugated paper to make a prototype of our light control device. It has one red light LED and the device is covered by several layers of black plastic bag, preventing any possibility of light leaking.
As mentioned above in the beginning, using red light as an input condition is an default environment in our project. Our ultimate goal is to regulate multiple genes as more as possible. So, in the future, by employing more different conditions, such as different lights, we would be able to regulate more genes.(one light sensor would result in controlling two more different genes) We hope that our project could eventually serve as a standardized model and set up with a standard operating procedure which offers future iGEMers to manipulate the control of multiple genes by simply changing the environmental conditions.
Reference
- Team : Imperial_College_London -2011.igem
- Team : Kyoto -2012.igem
 "
"