Team:UIUC Illinois/Project/Results
From 2013.igem.org
| (22 intermediate revisions not shown) | |||
| Line 41: | Line 41: | ||
<ul> | <ul> | ||
<li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Project/Overview">Overview</a></li> | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Project/Overview">Overview</a></li> | ||
| - | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Project/Background_Info">Background | + | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Project/Background_Info">Background</a></li> |
<li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Project/Design">Design</a></li> | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Project/Design">Design</a></li> | ||
<li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Project/Parts">Parts</a></li> | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Project/Parts">Parts</a></li> | ||
| Line 47: | Line 47: | ||
<li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:_UIUC_Illinois/Notebook">Notebook</a></li> | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:_UIUC_Illinois/Notebook">Notebook</a></li> | ||
</ul> | </ul> | ||
| - | <h3> | + | <h3>Achievements</h3> |
<ul> | <ul> | ||
<li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team_UIUC_Illinois/Medal_Requirements">Achievements</a></li> | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team_UIUC_Illinois/Medal_Requirements">Achievements</a></li> | ||
| Line 53: | Line 53: | ||
<h3>Human Practices</h3> | <h3>Human Practices</h3> | ||
<ul> | <ul> | ||
| - | |||
<li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Human_Practices/Outreach">Outreach</a></li> | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Human_Practices/Outreach">Outreach</a></li> | ||
<li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Human_Practices/Ethics">Ethics</a></li> | <li style="margin-bottom:20px;"><a href="https://2013.igem.org/Team:UIUC_Illinois/Human_Practices/Ethics">Ethics</a></li> | ||
| Line 66: | Line 65: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | |||
| + | |||
<div id="container" class="left"> | <div id="container" class="left"> | ||
<div id="major_image"> | <div id="major_image"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <img src="https://static.igem.org/mediawiki/2013/9/97/UIUC_Design-1.png" width="720" height="294" alt="Smiling fox" /> |
</div> | </div> | ||
| + | |||
| + | |||
<p class="left"> | <p class="left"> | ||
| - | Breakdown <br> <br> | + | <b>Breakdown</b> <br> <br> |
| - | As you may recall | + | As you may recall CDHcab is responsible for the breakdown of L-carnitine down our alternative pathway. We tested the function and production of cdh using a kinetic spectrophotometric analysis of NADH. As shown in the following reaction NADH is a product of when this enzyme converts L-carnitine into 3-dehydrocarnitine. Thus we can conclude whether or not our enzyme is working based on the levels of NADH accumulation after adding L-carnitine to a cytoplasmic extract containing our cdh enzyme.<br><br> |
| - | <img src="https:// | + | <p class="center"><img src="https://static.igem.org/mediawiki/2013/3/35/UIUC_Degradation.PNG" width="720" height="294" alt="degradation" /></p><br><br> |
| - | This was done by retrieving the cytoplasmic extract of our cells after induction of our CDH plasmid. This extract now containing the enzyme was | + | This was done by retrieving the cytoplasmic extract of our cells after induction of our CDH plasmid. This extract now containing the enzyme was analyzed for absorbance at 340 nm to watch NADH accumulation after adding either L-carnitine supspended in HEPES or HEPES alone. A spectrophotometer is able to give you this reading because NADH has specific absorbance wavelength that can be measured.<br><br> |
| - | By comparing the | + | <img src="https://static.igem.org/mediawiki/2013/6/6b/Carnitinegraphs1.PNG"><br><br> |
| - | + | By comparing the two graphs you can see that in comparison to the background accumulation of NADH that our cytoplasmic extract from the cell containing our genes accumulation of NADH is much greater. This indicates that our genes have created the correct enzyme since once L-carnitine is added the NADH level increases indicating that our intended reaction is proceeding. As depicted above the cytoplasmic extract from cells with the backbone alone did not generate an increase of NADH. This indicates that when L-carnitine was added to the cytoplasmic extract of the bacteria containing our gene the NADH concentration increased thus proving our desired enzyme is being produced and functioning as predicted. <br><br> | |
| + | |||
| + | <p class="center"><img src="https://static.igem.org/mediawiki/2013/1/1a/Carnitine3.PNG"><br><br></p> | ||
| + | |||
| + | This is a numerical representation of the collected data. Although the standard error between samples was higher than average, it still shows greater activity in the samples containing our gene of interest and L-carnitine than any other sample.<br><br> | ||
| + | |||
| + | |||
| + | <b>Uptake</b> <br> <br> | ||
| + | As you may recall the uptake system depends on two genes. CbcVW is a memebrane complex that allows L-carnitine to be shuttled through the membrane. CaiX is a free floating protein and directs the L-carnitine to our membrane protein thus helping the uptake of L-carnitine. We desired to measure the uptake of L-carnitine with Liquid Chromatography Mass Spectrometer. We will use this device by measuring the concentration of L-carnitine in the supernatant initially and finally. We prepared samples of Nissle with our genes and just with our backbone. Then we took samples of the supernatant initially and finally. By comparing L-carnitine concentrations before and after, the amount of L-carnitine that was uptaken will be visible. <br> | ||
| + | |||
| + | By looking at the graphs below you can see that our concentration of L-carnitine finally is drastically lower than the concentration of L-carnitine initially.<br> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/4/42/UIUC_Genes_lcms_intial.PNG" width="360" height="147" alt="lcms initial genes" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/2/24/UIUC_Genes_lcms_final.PNG" width="360" height="147" alt="lcms final genes" /> | ||
| + | </br> | ||
| + | It can also be seen that the bare backbone does not affect the concentration of L-carntine because initially and finally the concentration of L-carnitine is virtually identical. | ||
| + | <img src="https://static.igem.org/mediawiki/2013/b/b7/UIUC_Backbone_final_lcms.PNG" width="360" height="147" alt="lcms final backbone" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/9/93/UIUC_Lcms_inital_backbone.PNG" width="360" height="147" alt="lcms final backbone" /> | ||
| + | Conclusively it can be seen by this final overlaid graph of L-carnitine with our gene insert, that the concentration of L-carnitine in the supernatant drastically decreases over time. This indicates that our transporters are indeed accelerating the amount of L-carnitine that it uptakes. <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
</p> | </p> | ||
| + | <img alt="File:UIUC Comparison lcms pic new.PNG" src="/wiki/images/thumb/c/c8/UIUC_Comparison_lcms_pic_new.PNG/800px-UIUC_Comparison_lcms_pic_new.PNG" width="720" height="415"> | ||
</center> | </center> | ||
| + | |||
| + | </p> | ||
| + | <br><br> | ||
| + | <b>Deliver</b><br><br> | ||
| + | <p> | ||
| + | Day before:<p> | ||
| + | Nissle with streptomycin resistance was inoculated in lb+streptomycin. Also 1M calcium chloride and 1% sodium alginate by weight was autoclaved to sterilize and dissolve the sodium alginate.<br></p> | ||
| + | Day of: | ||
| + | <p> | ||
| + | E. coli Nissle was reinoculated in fresh 100ml LB and grown to an OD of 0.83. 1ml of the Nissle culture spun down at 13.4RPM for 1 min. Next 100μl of calcium chloride,100μl of sodium alginate, and 900μl of LB were added and the pellet of cells were resuspended. The mixture was then placed at 37˚C and shaken at 180RPM for ̴ 1hr. Next the mixture was spun down at 13.4 RPM for 1 min and the supernatant was poured out. The resulting pellet was a combination of Nissle and alginate gel. A control was also made of just an alginate pellet without Nissle added. These gels were transferred to gel capsules by Rocket Deals USA, bought from Amazon.com. To simulate how the pills would survive in the stomach we introduced the pills to HCl at a pH of 1 for 1 hour. Gel capsules are unable to withstand a pH of 1-2 so an enteric coating was put on the gel capsules by spraying non toxic Rust-Oleum Zinsser 408 Bulls Eye Clear Shellac spray bought from Amazon.com. The enteric coating allows the capsule to be stable at low pH's. After the hour of HCl treatment, the capsules were taken out and plated on a streptomycin lb agar plate. After 12 hours, growth on plates was evident as seen in figure 1. | ||
| + | </p><p> | ||
| + | <br>Figure 1: | ||
| + | <img style="-webkit-user-select: none; cursor: -webkit-zoom-in;" src="https://static.igem.org/mediawiki/2013/5/52/UIUC_Plate_growth.jpg" width="756" height="453"> | ||
| + | </p><p><br> | ||
| + | Two capsules with just bromothymol blue in them, one with an enteric coating and one without were tested. Bromothymol blue is a pH indicator that turns yellow at low pH's and blue at nuetral pH's (6-8). The capsule without the coating started breaking down in 5 minutes and was more or less completely disintegrated by time 15 minutes. This was clear by the change in color of HCl to yellow. The capsule with the coating did not break down for the full hour of testing. Figure 2 shows a comparison of the two capsules after the full hour.</p><p><br>Figure 2: | ||
| + | <img style="-webkit-user-select: none; cursor: -webkit-zoom-in;" src="https://static.igem.org/mediawiki/2013/c/c8/UIUC_comparisonofph1.jpg" width="756" height="453"></p><p><br> | ||
| + | After the hour, the capsule with enteric coating was introduced to water at a pH close to 7. Here the capsule started to break down, turning the solution blue. These results show that the probiotic is able to survive the stomach and target the small intestine. | ||
| + | <p><br> | ||
| + | Figure 3: | ||
| + | <img style="-webkit-user-select: none; cursor: -webkit-zoom-in;" src="https://static.igem.org/mediawiki/2013/2/29/UIUC_ph7_entericcoating.jpg" width="728" height="453"></p> | ||
| + | |||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 03:57, 29 October 2013

Breakdown
As you may recall CDHcab is responsible for the breakdown of L-carnitine down our alternative pathway. We tested the function and production of cdh using a kinetic spectrophotometric analysis of NADH. As shown in the following reaction NADH is a product of when this enzyme converts L-carnitine into 3-dehydrocarnitine. Thus we can conclude whether or not our enzyme is working based on the levels of NADH accumulation after adding L-carnitine to a cytoplasmic extract containing our cdh enzyme.
This was done by retrieving the cytoplasmic extract of our cells after induction of our CDH plasmid. This extract now containing the enzyme was analyzed for absorbance at 340 nm to watch NADH accumulation after adding either L-carnitine supspended in HEPES or HEPES alone. A spectrophotometer is able to give you this reading because NADH has specific absorbance wavelength that can be measured.
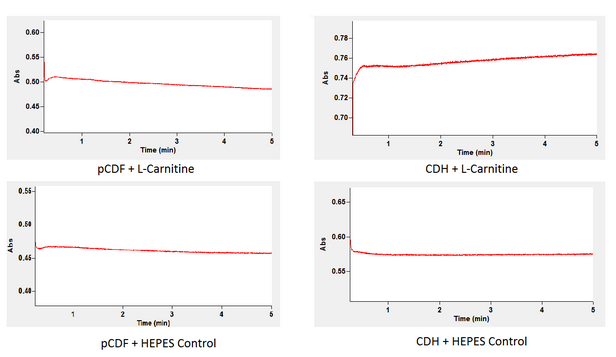
By comparing the two graphs you can see that in comparison to the background accumulation of NADH that our cytoplasmic extract from the cell containing our genes accumulation of NADH is much greater. This indicates that our genes have created the correct enzyme since once L-carnitine is added the NADH level increases indicating that our intended reaction is proceeding. As depicted above the cytoplasmic extract from cells with the backbone alone did not generate an increase of NADH. This indicates that when L-carnitine was added to the cytoplasmic extract of the bacteria containing our gene the NADH concentration increased thus proving our desired enzyme is being produced and functioning as predicted.
Uptake
As you may recall the uptake system depends on two genes. CbcVW is a memebrane complex that allows L-carnitine to be shuttled through the membrane. CaiX is a free floating protein and directs the L-carnitine to our membrane protein thus helping the uptake of L-carnitine. We desired to measure the uptake of L-carnitine with Liquid Chromatography Mass Spectrometer. We will use this device by measuring the concentration of L-carnitine in the supernatant initially and finally. We prepared samples of Nissle with our genes and just with our backbone. Then we took samples of the supernatant initially and finally. By comparing L-carnitine concentrations before and after, the amount of L-carnitine that was uptaken will be visible.
By looking at the graphs below you can see that our concentration of L-carnitine finally is drastically lower than the concentration of L-carnitine initially.

 It can also be seen that the bare backbone does not affect the concentration of L-carntine because initially and finally the concentration of L-carnitine is virtually identical.
It can also be seen that the bare backbone does not affect the concentration of L-carntine because initially and finally the concentration of L-carnitine is virtually identical.

 Conclusively it can be seen by this final overlaid graph of L-carnitine with our gene insert, that the concentration of L-carnitine in the supernatant drastically decreases over time. This indicates that our transporters are indeed accelerating the amount of L-carnitine that it uptakes.
Conclusively it can be seen by this final overlaid graph of L-carnitine with our gene insert, that the concentration of L-carnitine in the supernatant drastically decreases over time. This indicates that our transporters are indeed accelerating the amount of L-carnitine that it uptakes. 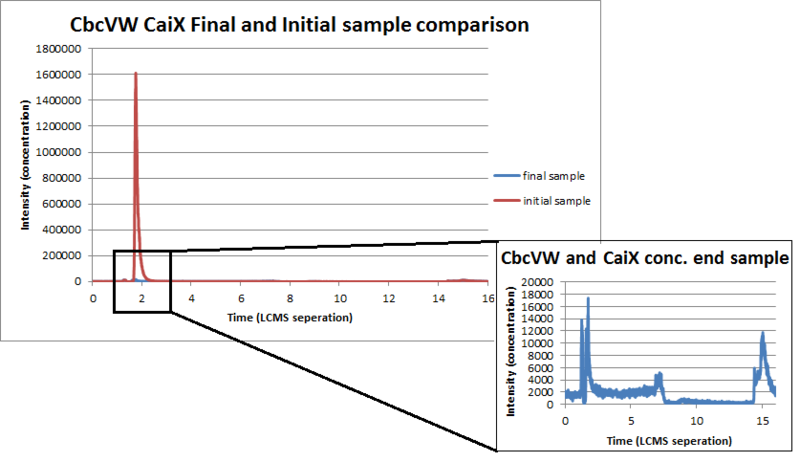
Deliver
Day before:
Nissle with streptomycin resistance was inoculated in lb+streptomycin. Also 1M calcium chloride and 1% sodium alginate by weight was autoclaved to sterilize and dissolve the sodium alginate.
E. coli Nissle was reinoculated in fresh 100ml LB and grown to an OD of 0.83. 1ml of the Nissle culture spun down at 13.4RPM for 1 min. Next 100μl of calcium chloride,100μl of sodium alginate, and 900μl of LB were added and the pellet of cells were resuspended. The mixture was then placed at 37˚C and shaken at 180RPM for ̴ 1hr. Next the mixture was spun down at 13.4 RPM for 1 min and the supernatant was poured out. The resulting pellet was a combination of Nissle and alginate gel. A control was also made of just an alginate pellet without Nissle added. These gels were transferred to gel capsules by Rocket Deals USA, bought from Amazon.com. To simulate how the pills would survive in the stomach we introduced the pills to HCl at a pH of 1 for 1 hour. Gel capsules are unable to withstand a pH of 1-2 so an enteric coating was put on the gel capsules by spraying non toxic Rust-Oleum Zinsser 408 Bulls Eye Clear Shellac spray bought from Amazon.com. The enteric coating allows the capsule to be stable at low pH's. After the hour of HCl treatment, the capsules were taken out and plated on a streptomycin lb agar plate. After 12 hours, growth on plates was evident as seen in figure 1.
Figure 1:

Two capsules with just bromothymol blue in them, one with an enteric coating and one without were tested. Bromothymol blue is a pH indicator that turns yellow at low pH's and blue at nuetral pH's (6-8). The capsule without the coating started breaking down in 5 minutes and was more or less completely disintegrated by time 15 minutes. This was clear by the change in color of HCl to yellow. The capsule with the coating did not break down for the full hour of testing. Figure 2 shows a comparison of the two capsules after the full hour.
Figure 2:

After the hour, the capsule with enteric coating was introduced to water at a pH close to 7. Here the capsule started to break down, turning the solution blue. These results show that the probiotic is able to survive the stomach and target the small intestine.
Figure 3:

 "
"



