Team:XMU-China/Content3
From 2013.igem.org
| (27 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<html> | <html> | ||
<head> | <head> | ||
| - | <meta content="text/html; charset=utf-8 | + | <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> |
<title>LinkUp - Multipurpose HTML Template</title> | <title>LinkUp - Multipurpose HTML Template</title> | ||
<link rel="icon" type="image/png" href="images/favicon.png" /> | <link rel="icon" type="image/png" href="images/favicon.png" /> | ||
| + | <style> | ||
| + | #wrapper p{ font-size:19px;} | ||
| + | #wrapper1 p{ font-size:19px;} | ||
| + | #wrapper2 p{ font-size:19px;} | ||
| + | table{font-size:12px;background-color:transparent;text-align:center;} | ||
| + | </style> | ||
<!--[if lt IE 9]> | <!--[if lt IE 9]> | ||
<link rel="stylesheet" type="text/css" href="css/ie-fix.css" /> | <link rel="stylesheet" type="text/css" href="css/ie-fix.css" /> | ||
<![endif]--> | <![endif]--> | ||
<link rel="stylesheet" type="text/css" href="https://2013.igem.org/Team:XMU-China/reset.css?action=raw&ctype=text/css" > | <link rel="stylesheet" type="text/css" href="https://2013.igem.org/Team:XMU-China/reset.css?action=raw&ctype=text/css" > | ||
| - | <link rel="stylesheet" | + | <link rel="stylesheet" typece="text/css" href="https://2013.igem.org/Team:XMU-China/style2.css?action=raw&ctype=text/css" > |
<link rel='stylesheet' type='text/css' href='http://fonts.googleapis.com/css?family=Oswald' > | <link rel='stylesheet' type='text/css' href='http://fonts.googleapis.com/css?family=Oswald' > | ||
| Line 22: | Line 28: | ||
<style> | <style> | ||
#wrapper {background:#fffccc} | #wrapper {background:#fffccc} | ||
| + | |||
</style> | </style> | ||
| - | + | <div id="separator"> | |
| - | + | <div class="center-block"> | |
<a name="Background"></a> | <a name="Background"></a> | ||
| - | + | <h3>Background</h3> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <!-- START CONTENT --> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<div class="center-block-page clearfix"> | <div class="center-block-page clearfix"> | ||
| - | + | ||
| - | + | <h4>Forming the Glee</h4> | |
| - | + | <p>To be a good glee, first it needs a good song. The most intriguing song on earth is called OSCILLATION. If you look about, you will find it ubiquitous and extremely useful. After we taught each member this song, we need to help them sing it together, so oscillation became SYNCHRONIZED OSCILLATION. Thus how we form a glee! | |
| - | The | + | </p> |
| - | + | ||
| - | + | <h4>Oscillation: The Magic Song of World</h4> | |
| - | + | <p>Once upon the time, there was a magic song called oscillation, it permeated every corner of the world. It <b>regulated nearly everything</b> on the earth. The interaction of moon and ocean generated the magnificent tide oscillation, and the biological process that displays a 24-hour oscillation inside human body.</p> | |
| - | < | + | <table ><tr><td><img src="https://static.igem.org/mediawiki/2013/7/75/Fig1_1.jpg" width =308px height=286px class="border" alt="" /> </td> |
| - | + | <td><img src="https://static.igem.org/mediawiki/2013/5/5f/Fig1_2.png" width =554px height=286px class="border" alt="" /> | |
| - | + | </td></tr> | |
| - | + | <tr><td>Fig. 1-1 The Tides depend on several factors, including</br> where the sun and moon are relative to the Earth</td><td>Fig. 1-2 The circadian clock inside human body</td></tr></table></br></br> | |
| - | + | <p> | |
| + | Besides, oscillation also <b>plays an important part </b>in signal transition. Take lasers for example, which are known for their intensity and can be focused to a tight spot over long distance. These characteristics all owe to their spatial coherent in the frequency of the light source. | ||
</p> | </p> | ||
| - | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/1/1c/Xmubg-Image003.jpg" width =308px height=286px class="border" alt="" /> </td> | |
| - | + | </tr> | |
| - | + | <tr><td>Fig. 1-3 Helium-Neon laser demonstration at the</br> Kastler-Brossel Laboratory at Paris VI: Pierre et Marie</td></table></br></br> | |
| - | + | <h4>Oscillation in Bacteria</h4> | |
| - | + | <p> | |
| - | </ | + | Synthetic biologists have done a lot of work to teach bacteria to sing this oscillation song by building artificial genetic circuit inside them, since oscillations can lead to fantastic applications and benefit our everyday life. Besides regulation and signal transmission, oscillations in living organisms can also react to its growing environment, which probably will activate the trigger of oscillation inside cells and generate oscillation signals. From collected signals we can tell how an environment factor influences the cells behavior, and in turn the environment factor can be indicated by the signals. Furthermore, if we interpret these kinds of oscillation signals like 1010 in binary system, then we could even make an electronic environmental factor detector, using signals from environment as an input, and numbers on computer screen as output (Fig. 1-4). |
| - | < | + | </p> |
| - | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/f/f9/Fig1_4.png" width =500px height=262px class="border" alt="" /> </td> | |
| - | < | + | </tr> |
| - | <br | + | <tr><td>Fig. 1-4 How to make a signal converter</td></table></br></br> |
| + | |||
| + | <p>Here is the progress of artificial synthetic biological oscillations have made.<br/> | ||
| + | In <b>the first generation of oscillation</b>, three transcriptional repressor systems that are not part of any natural biological clock are used to build an oscillating network, termed the repressilator, in <i>Escherichia coli</i> (Fig. 1-5). The network periodically induces the synthesis of green fluorescent protein as a readout of its state in individual cells. However, plasmids can hardly pass from generation to generation and this artificial clock is not robust enough and always displays noisy behavior. | ||
| + | </p> | ||
| + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/1/13/Xmu-bgImage007.png" class="border" alt="" /> </td> | ||
| + | </tr> | ||
| + | <tr><td >Fig. 1-5 The first generation oscillator, known as repressor</td></tr></table></br></br> | ||
| + | |||
| + | <p>Scientists constructed the second generation oscillator rapidly. This <b>improved oscillator</b> is fast, robust and persistent; with tunable oscillatory periods as fast as 13 min (Fig.1-6). The oscillator is designed using a previously modeled network architecture comprising linked positive and negative feedback loops. But this circuit could only monitor oscillations in individual cells through multiple cycles, which means the song is still limited to single individual. | ||
| + | </p> | ||
| + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/e/eb/Xmu-bgImage009.png" class="border" alt="" /> </td> | ||
| + | </tr> | ||
| + | <tr><td>Fig. 1-6 2nd generation of oscillation consists of positive and negative feedback loops</td></table></br></br> | ||
| + | |||
| - | < | + | <p>Thinking that singing alone is lonely, scientists introduce quorum sensing to connect isolated cells and let them sing together, and technically it's called synchronized oscillation. The synchronization effect of quorum sensing, however, is limited to over tens of micrometer (Fig.1-7). If the synchronized colony could be enlarged, then the oscillation signal will be much stronger and steadier. Longer ranged synchronized oscillation is realized by a gas-phase redox signal H2O2 to the third generation circuit, which could achieve the communication among colonies at over millimeter scales and became the fourth generation. Oh yeah, finally our bacteria can sing the same oscillation at the same time, so they decided to form their own glee! |
| - | + | </p> | |
| - | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/c/c9/Xmu-bgImage012.png" class="border" alt="" /> </td> | |
| - | + | </tr> | |
| - | + | <tr><td>Fig.1-7 3rd generation involved in quorum sensing</td></table></br></br> | |
| - | + | <p>That's how our little <i>E. coli</i> friend formed their glee, a long but cheerful story. Thanks to hardworking synthetic biologists. Let's go and see how this fourth generation oscillation glee functions inside in <b>THREE PLASMIDS</b>!</p> | |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
<p><i>Reference<br/></i> | <p><i>Reference<br/></i> | ||
1. http://en.wikipedia.org/wiki/Oscillation<br/> | 1. http://en.wikipedia.org/wiki/Oscillation<br/> | ||
| Line 92: | Line 90: | ||
3. Stricker J. et al. A fast, robust and tunable synthetic gene oscillator. <i>Nature</i> 456, 516-519 (2008)<br/> | 3. Stricker J. et al. A fast, robust and tunable synthetic gene oscillator. <i>Nature</i> 456, 516-519 (2008)<br/> | ||
4. Danino T., Mondrago´n-Palomino O., Tsimring L. & Hasty J. A synchronized quorum of genetic clocks. <i>Nature</i> 463, 326-330 (2010)<br/> | 4. Danino T., Mondrago´n-Palomino O., Tsimring L. & Hasty J. A synchronized quorum of genetic clocks. <i>Nature</i> 463, 326-330 (2010)<br/> | ||
| - | </p> | + | </p> </div><!-- .center-block-page --> |
| - | + | ||
| - | + | <!-- END CONTENT --> | |
</div><!-- #wrapper --> | </div><!-- #wrapper --> | ||
| - | <!-- END WRAPPER --> | + | <!-- END WRAPPER --> |
<div id="wrapper1"> | <div id="wrapper1"> | ||
| Line 104: | Line 101: | ||
#wrapper1 {background:#cfffcd} | #wrapper1 {background:#cfffcd} | ||
</style> | </style> | ||
| - | + | <div id="separator"> | |
| - | + | <div class="center-block"> | |
<a name="Mechanism"></a> | <a name="Mechanism"></a> | ||
| - | + | <h3>Mechanism</h3> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <!-- START CONTENT --> | |
| - | + | <div class="center-block-page clearfix"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <h4>Three Parts in Circuit Designation</h4> <br/> |
| - | + | <p> | |
| - | <div class=" | + | In a synchronized oscillatory system, three important parts should be included: the <b>oscillator</b>, which is the biochemical machinery that generate the oscillatory output; the <b>coupling</b> pathway that ensure the connection among cells; and output pathway, which is also known as a <b>reporter</b> that reflect the state of the oscillator to downstream targets (Fig. 2-1). |
| - | + | </p> | |
| - | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/f/fe/Xmum-Image001.png" class="border" alt="" /> </td> | |
| - | + | <td><img src="https://static.igem.org/mediawiki/2013/b/ba/Fig2_1.jpg" class="border" alt="" /> | |
| - | + | </td></tr> | |
| - | + | <tr><td>Fig. 2-1 Circuit working mechanism</td></table> | |
| - | + | ||
| - | + | ||
| - | + | </div><!-- .center-block-page --> | |
| + | <div class="center-block-page clearfix"> | ||
| + | |||
| + | <h4>Oscillator</h4> | ||
| + | |||
| + | <p> | ||
| + | Quorum sensing (QS) is a cell-to-cell signaling mechanism that refers to the ability of bacteria to respond to chemical hormone-like molecules called autoinducers. When an autoinducer reaches a critical threshold, the bacteria detect and respond to this signal by altering their gene expression. In our circuit, QS (from <i>Vibrio fischeri</i>) is installed as a positive feedback while <i>aiiA</i> (from <i>Bacillus Thurigensis</i>) acts as a negative one to compose an oscillator together. In the quorum sensing part, the <i>luxI</i> gene is at low expression level and produces LuxI protein that synthesize a kind of acyl-homoserine lactone (AHL), which is a small molecule that can diffuse across the cell membrane and mediate intercellular coupling when it reaches the threshold as enough biomass accumulated. AHL will bind intracellular protein LuxR, which is also consecutively produced by <i>luxR</i> gene. The LuxR-AHL complex can activate the <i>luxI</i> promoter, and the positive feedback loop is built. At the same time, the <i>aiiA</i> gene, which is under control of the same promoter is expressed and produce a lactonase enzyme known as <i>aiiA</i> that hydrolyzes the lactone ring of AHL. (Fig. 2-1) In this system, the activator enhances the expression of both activator and repressor, which shares the common motif of many synthetic oscillators. | ||
| + | </p> | ||
| + | </div><!-- .center-block-page --> | ||
| + | <div class="center-block-page clearfix"> | ||
| + | <h4>Coupling</h4> | ||
| + | <p> | ||
| + | The communication range of quorum sensing, however, is limited by the diffusion rate of AHL, could only reach cells over tens of micrometers. So, we introduced the coding gene for NADH dehydrogenase II (<i>ndh</i>) and put it under the control of the same <i>luxI</i>promoter, which means <i>ndh</i> also has a periodical expression in accordance with the oscillator. NDH-2 is a membrane-bound respiratory enzyme that generates low level H<sub>2</sub>O<sub>2</sub> and superoxide (O<sub>2</sub><sup>-</sup>) and H<sub>2</sub>O<sub>2</sub> will permeate to neighboring colonies. Periodic production of H<sub>2</sub>O<sub>2</sub> changes the redox state of a cell immediately and interacts with the synthetic circuit through the native aerobic response control systems, including ArcAB, which has a binding site in the <i>lux</i> promoter region. Before the oxidizing condition is triggered, ArcAB is partially expressed, so <i>lux</i> is partially repressed. When the concentration of H<sub>2</sub>O<sub>2</sub> is increasing, ArcAB is gradually inactivated, and the repression of <i>lux</i> is relieved. With the periodically produced vapor phase H<sub>2</sub>O<sub>2</sub> that can diffuse quickly among colonies, the oscillation is not only strengthened but also expanded to millimeter scales(Fig. 2-2).<br/><br/></p | ||
| + | > <table><tr><td><img src="https://static.igem.org/mediawiki/2013/8/88/ArCab.png" width=500 height=300 class="border" alt="" /> </td> | ||
| + | </tr> | ||
| + | <tr><td>Fig. 2-2 H<sub>2</sub>O<sub>2</sub> functions as an activator to the quorum sensing promoter</td></table> | ||
| - | + | <p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p> | + | |
When compared with the strong but short ranged coupling by QS, H<sub>2</sub>O<sub>2</sub> might be weaker but long ranged because its disperse characteristic. These two levels of communication between cells formed the basic oscillatory system inside our host. | When compared with the strong but short ranged coupling by QS, H<sub>2</sub>O<sub>2</sub> might be weaker but long ranged because its disperse characteristic. These two levels of communication between cells formed the basic oscillatory system inside our host. | ||
| - | + | </p> | |
| - | + | </div><!-- .center-block-page --> | |
| - | + | <div class="center-block-page clearfix"> | |
| - | + | <h4>Reporter</h4> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<p> | <p> | ||
| - | + | A good reporter in an oscillatory circuit should be steadily expressed and can be easily detected by regular equipment in laboratory, so broadly used reporter green fluorescent protein (GFP) came into our mind.<br/><br/> | |
| - | + | First, we built regular <i>gfp</i> gene into our circuit and it worked well. Its fluorescence can be observed by fluorescence microscopy under 1s of exposure time on the microfluidic array 1(Fig. 2-3).</p> | |
| - | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/b/b4/Xmum-Image003.jpg" class="border" alt="" /> </td> | |
| - | + | </tr> | |
| - | + | <tr><td>Fig. 2-3 GFP under fluorescence microscopy on a microfluidic array</td></table></br> | |
| - | < | + | |
| - | + | ||
<p> | <p> | ||
| - | P. S. | + | However, there is a newly found GFP variant Superfolder GFP, which has shown outstanding performance in many ways. It has two advantages in observing: |
| - | Please come with us to see how we built our magic circuit in E.coli in Construction!</p> | + | </br>1) Fold 3.5 to 4 times faster than regular GFP; </br> |
| - | + | 2) Yield up to four times more total Fluorescence than regular GFP.<br/> | |
| - | + | The greater the fluorescence strength is, the shorter exposure time will be needed, thus can decrease the photobleaching (GFP will be eventually destroyed by the light exposure that tries to stimulate it into fluorescing) that will lead into lapses in data processing.<br/> | |
| - | + | Besides, we also want to make a comparison between two different GFPs' performances in our oscillatory circuit. For example, peaks, troughs and periods in oscillatory curves. The following picture shows the difference between GFP and sfGFP both on colE1 backbone.(Fig. 2-4)</p> | |
| + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/4/4c/Xmum-Image004.jpg" width=500 height=332 class="border" alt="" /> </td> | ||
| + | </tr> | ||
| + | <tr><td>Fig.2-4 Difference between GFP and sfGFP both on colE1 backbone</br> | ||
| + | (P. S. Thanks the Peking iGEM team 2013 for providing us the sfGFP part.) | ||
| + | </td></table> | ||
| + | |||
| + | <p> | ||
| + | Please come with us to see how we built our magic circuit in <i>E. coli</i> in Construction!</p> | ||
| + | |||
| + | |||
| + | </div><!-- .center-block-page --> | ||
| - | + | <!-- END CONTENT --> | |
</div><!-- #wrapper --> | </div><!-- #wrapper --> | ||
| - | <!-- END WRAPPER --> | + | <!-- END WRAPPER --> |
<div id="wrapper2"> | <div id="wrapper2"> | ||
| Line 179: | Line 179: | ||
#wrapper2 {background:#ffdcb2} | #wrapper2 {background:#ffdcb2} | ||
</style> | </style> | ||
| - | + | <div id="separator"> | |
| - | + | <div class="center-block"> | |
<a name="Circuit_Construction"></a> | <a name="Circuit_Construction"></a> | ||
| - | + | <h3>Circuit Construction</h3> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <!-- START CONTENT --> | |
| - | + | <div class="center-block-page clearfix"> | |
<h4>Overview</h4> | <h4>Overview</h4> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<p> | <p> | ||
| - | + | So we designed three plasmids to function as the three important parts in the oscillation circuit, and all of them are in the charge of the same quorum sensing promoter to make sure that all target genes share the same oscillatory period (Table 2-1). | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</p> | </p> | ||
| - | + | <table width="100%" border="1" cellspacing="0px" align="center"> | |
| - | + | <caption> | |
| - | + | Table 2-1 Three plasmids constructed as parts of our circuit | |
| - | + | </caption> | |
| - | + | <tr> | |
| - | <table | + | <th rowspan=2>No.</th> |
| - | + | <td rowspan=2>Plasmid</td> | |
| - | + | <td rowspan=2>Replication</br>Origin</td> | |
| - | + | <td rowspan=2>Copy</br>Number</td> | |
| - | + | <td rowspan=2>Resistance</td> | |
| - | + | <td colspan=2 >Size(bp)</td> | |
| - | + | ||
| - | + | </tr> | |
| - | + | <tr> | |
| - | < | + | <td>Insert</td> |
| - | + | <td>Backbone</td> | |
| - | + | ||
| - | + | </tr> | |
| - | + | <tr> | |
| - | + | <th >A1</th> | |
| - | + | <td>pSB1C3-gfp-luxI</td> | |
| - | + | <td>pSB1C3</td> | |
| - | + | <td>high</br>(100~300)</td> | |
| - | + | <td>Chloromycetin</br>(Cm)</td> | |
| - | + | <td>4052</td> | |
| - | + | <td>2070)</td> | |
| - | + | </tr> | |
| - | + | <tr> | |
| - | + | <th >A2</th> | |
| - | </ | + | <td>pSB1C3-sfgfp-luxI</td> |
| - | < | + | <td>pSB1C3</td> |
| - | + | <td>high</br>(100~300)</td> | |
| - | + | <td>Chloromycetin</br>(Cm)</td> | |
| - | + | <td>4046</td> | |
| - | + | <td>2070)</td> | |
| - | + | </tr> | |
| - | + | <tr> | |
| - | + | <th >A3</th> | |
| - | + | <td>p3H-GFP-luxI</td> | |
| - | < | + | <td>p3H</td> |
| - | + | <td>Middle</br>(18~22)</td> | |
| - | < | + | <td>Ampicillin</br> |
| - | + | (Amp)</td> | |
| - | + | <td>4052</td> | |
| - | + | <td>/</td> | |
| - | + | </tr> | |
| - | + | <tr> | |
| - | + | <th >A4</th> | |
| - | + | <td>p3H-sfGFP-luxI</td> | |
| - | + | <td>p3H</td> | |
| - | + | <td>Middle</br>(18~22)</td> | |
| - | + | <td>Ampicillin</br> | |
| - | + | (Amp)</td> | |
| - | + | <td>4046</td> | |
| - | + | <td>/</td> | |
| - | + | </tr> | |
| - | + | <tr> | |
| - | + | <th >B</th> | |
| - | + | <td>pSB3T5-aiiA</td> | |
| - | + | <td>p15A</td> | |
| - | + | <td>Middle</br>(10~12)</td> | |
| - | + | <td>Tetracycline</br> | |
| - | + | (Tet) | |
| - | + | </td> | |
| - | + | <td>2135</td> | |
| - | + | <td>2837</td> | |
| - | + | </tr> | |
| - | + | ||
| - | + | <tr> | |
| - | + | <th >C</th> | |
| - | + | <td>pSB4K5-ndh</td> | |
| - | + | <td>pSC101</td> | |
| - | + | <td>Low</br>(5)</td> | |
| - | + | <td>Kanamycin</br> | |
| - | + | (Kan) | |
| - | + | </td> | |
| - | + | <td>2658</td> | |
| - | + | <td>3004</td> | |
| - | + | </tr> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</table> | </table> | ||
| - | |||
| + | </br> | ||
| - | < | + | <p>In a synchronized oscillation circuit, each A, B and C plasmid is necessary. The plasmid A expresses GFP <b>breporter</b> and LuxI proteins, which is the <b>positive feedback in oscillator</b>; the plasmid B expresses protein aiiA to degrade AHL and acts as the<b> negative feedback in oscillator</b>; the plasmid C expresses NAD-2 to generate H2O2 to communicate between colonies, is the <b>coupling part</b> in circuit (Fig 2-5). </p> |
| - | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/0/06/Fig2_5.jpg" class="border" alt="" /> </td> | |
| - | + | </tr> | |
| - | + | <tr><td>Fig. 2-5 Plasmids A, B and C in oscillation circuit</td></table></br> | |
| - | + | <p>You may have noticed that these three plasmids have different resistant genes and replication origins on their backbone. Different resistant genes are used mainly for selection. And since different replication origin means a different copy number of target genes in this plasmid. Using different replication origins is to produce different target proteins in an appropriate ratio, and only in this way can oscillation be observed. Just like a glee needs Soprano, Tenor and Bass to cooperate to finish a song. Besides, three artificial plasmids with same copy numbers always have a competitive relation in one cell and cannot be well expressed.</p> | |
| - | + | <p>Plasmid A has two different backbones with high and middle copy numbers respectively. We built them to see the copy numbers' effect on oscillation. Results can be seen in <b>Improvement (For a better Glee)</b>. </p> | |
| - | + | <p>All of the plasmids we constructed are confirmed by agarose gel electrophoresis, and result can be seen in <b>Parts</b>, where you can also get the link to parts we submitted. </p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p> | + | <p>After the onerous construction work, our glee is ready to perform the synchronized oscillation song, so we have to find them a stage. Let's go to see the beautiful opera in <b>Microfluidics (Stages)</b>. </p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p><i>Reference<br/></i> | |
| - | + | 1. Prindle, A., et al., A sensing array of radically coupled genetic 'biopixels'. <i>Nature</i> 481, 39-44, 2012;<br/> | |
| - | + | 2. Pedelacq, J. et al., Engineering and characterization of a superfolder green fluorescent protein. <i>Nature 24</i>, 79-88, 2006;<br/> | |
| - | + | 3. Waters C. M. & Bassler B. L. Quorum Sensing: Cell-to-Cell Communication in <i>Bacteria. Annu. Rev. Cell Dev. Biol</i> 21, 319-46, 2005.<br/> | |
| - | + | </p> | |
| + | </div></div> | ||
| + | <!-- .center-block-page --> | ||
| - | + | <!-- END CONTENT --> | |
</div><!-- #wrapper --> | </div><!-- #wrapper --> | ||
| - | <!-- END WRAPPER --> | + | <!-- END WRAPPER --> |
<div id="wrapper"> | <div id="wrapper"> | ||
| - | + | <div id="separator"> | |
| - | + | ||
| - | + | <div class="center-block" style:"width: 1000px;"> | |
<a name="Microfluidic"></a> | <a name="Microfluidic"></a> | ||
| - | + | <h3>MICROFLUIDIC & MICROSCOPY</h3> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <!-- START CONTENT --> | |
| - | + | <div class="center-block-page clearfix"> | |
| - | <h4> | + | <h4>Microfluidics (Opera Houses)</h4> |
| - | < | + | <p>A Microfluidic array to cells is like an opera house to a glee that provides a cell culture environment and observation stage. To accompany the observation of microfluidic we used the fluorescence microscope, just like mass media to photograph the oscillation. We call them M & M. What a lovely couple.</p> |
| - | + | ||
| - | + | <h4>Why M&M?</h4> | |
| - | + | ||
| - | + | <p> | |
If we want to capture gene expression dynamics and distributions reported by fluorescence, we need to create a chemostat that provide a near-constant environment for the growth of bacteria. The near-constant environment mainly includes two aspects: continuous fresh media to ensure the optimum growth state and constant small populations of bacteria to ensure single-cell resolution in observation. Steady growth state is needed because the oscillation curve is expected to be a near simple harmonic one. Besides, bacteria tend to form biofilm which will lead to the overlapping of cells and create a false impression that the fluorescence increases, and that's why we have to prevent this to maintain a constant bacteria population with single-cell resolution.<br/><br/> | If we want to capture gene expression dynamics and distributions reported by fluorescence, we need to create a chemostat that provide a near-constant environment for the growth of bacteria. The near-constant environment mainly includes two aspects: continuous fresh media to ensure the optimum growth state and constant small populations of bacteria to ensure single-cell resolution in observation. Steady growth state is needed because the oscillation curve is expected to be a near simple harmonic one. Besides, bacteria tend to form biofilm which will lead to the overlapping of cells and create a false impression that the fluorescence increases, and that's why we have to prevent this to maintain a constant bacteria population with single-cell resolution.<br/><br/> | ||
What we mentioned above might have covered all aspects we need to consider for communication in a single colony, but if we still want to observe the coupling between colonies we have to create an array that contains many colonies and allows H<sub>2</sub>O<sub>2</sub> to pass through freely.<br/><br/> | What we mentioned above might have covered all aspects we need to consider for communication in a single colony, but if we still want to observe the coupling between colonies we have to create an array that contains many colonies and allows H<sub>2</sub>O<sub>2</sub> to pass through freely.<br/><br/> | ||
| - | Among many different techniques, microfluidic array seems to be our best choice with following advantages:< | + | Among many different techniques, microfluidic array seems to be our best choice with following advantages:</br> |
1) Precisely control of flowing rate and cell populations when equipped with syringe pumps;<br/> | 1) Precisely control of flowing rate and cell populations when equipped with syringe pumps;<br/> | ||
2) Cells behavior including fluorescence strength can be captured by high resolution microscopy;<br/> | 2) Cells behavior including fluorescence strength can be captured by high resolution microscopy;<br/> | ||
| Line 781: | Line 328: | ||
| - | + | </p> | |
| - | + | <div class="clear"></div> | |
| - | + | <div class="clear"></div> | |
| - | + | ||
| - | + | </div><!-- .center-block-page --> | |
| - | + | ||
<div class="center-block-page clearfix"> | <div class="center-block-page clearfix"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <h4>Design of Microfluidic Array</h4> | ||
| + | <p><b>1.Pretests in "Swimming Pool"</b></p> | ||
| + | <p>Before our experiments on the microfluidic array we described before, a series of pretests were done on a chip whose structure looks like many swimming pool on the ground (Fig. 3-1).</p> | ||
| + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/6/6b/Xmumm-Image012.png" width=500 height=300 class="border" alt="" /> </td> | ||
| + | </tr> | ||
| + | <tr><td>Fig. 3-1 A "Swimming pool" chip in black field; those white squares (100μm×100μm×100μm) are chambers for bacteria</td></table></br> | ||
| + | <p> | ||
| + | From the sketch (Fig. 3-2) we can see that the chamber is like a swimming pool where bacteria swim happily in the fresh LB that running from up above, and newly generated cells would be washed away. | ||
| + | </p> | ||
| + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/5/5d/Xmumm-Image014.png" width=700 height=290 class="border" alt="" /> </td> | ||
| + | </tr> | ||
| + | <tr><td>Fig. 3-2 Diagram for swimming pool model's working mechanism</td></table></br> | ||
| - | According to what we have discussed above, the microfluidic device should be able to provide flowing fresh media and maintain a constant bacteria population with single-cell resolution. | + | <p>A "swimming pool" chamber model would have a lager AHL concentration when compared with a "passage" chamber because the former one has a minor dilution. According to a published research that the oscillation period will be shorter as the AHL concentration increases. So we assume that it will be more time-consuming to use this "swimming pool" model in pretests.</p> |
| - | + | <p>However, this chamber is about 100 μm deep and could grow E. coli far more than monolayer. As time goes by, the oscillation in a single colony signaling by AHL could never go back to the original baseline, and the oscillation curves have a tendency like the following graph (Fig. 3-3).</p> | |
| - | <b>Channels and Chambers</b> | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/a/af/Xmumm-Image016.png" width=500 height=300 class="border" alt="" /> </td> |
| - | + | </tr> | |
| + | <tr><td>Fig. 3-3 Simulation curve for our oscillation in "Swimming Pool"</td></table></br> | ||
| + | |||
| + | |||
| + | <p><b>2.Advanced "Passage"</b></p> | ||
| + | <p>According to what we have discussed above, the microfluidic device should be able to provide flowing fresh media and maintain a constant bacteria population with single-cell resolution. Because "swimming pool" has a cell stacking problem, we designed a new passage-like device as shown below (Video 1).</p> | ||
| + | |||
| + | <h4>Video 1</h4> | ||
| + | <center> | ||
| + | <iframe width="640" height="360" src="http://www.youtube.com/embed/enLFOdr5P-M?feature=player_detailpage" frameborder="0" allowfullscreen></iframe><br/></center> | ||
| + | |||
| + | <p> | ||
| + | <b>Channels and Chambers</b> </p> | ||
| + | <p> | ||
This device is composed by two parts: the flowing channels and near 3,000 square trapping chambers, and their layout is like there are many passages with symmetry rooms distributed on both sides, so we call this array "passage" model. Firstly, cells are loaded from the cell port. When the loading stops, most cells would remain in channels, but if the loading cell density is dense enough, many cells would be pushed into chambers and form a monolayer of Escherichia coli growing in it. This is the time for running LB to get in. Fresh LB is loaded from the media port with Tween 20* and appropriate antibiotics. Flowing LB does not only permit a constant nutrition and exponentially growing cells for more than two days (that's the longest time we have tried), but also washes away newly generated cells to sustain a certain cell density, which is crucial for the determination of fluorescence strength. | This device is composed by two parts: the flowing channels and near 3,000 square trapping chambers, and their layout is like there are many passages with symmetry rooms distributed on both sides, so we call this array "passage" model. Firstly, cells are loaded from the cell port. When the loading stops, most cells would remain in channels, but if the loading cell density is dense enough, many cells would be pushed into chambers and form a monolayer of Escherichia coli growing in it. This is the time for running LB to get in. Fresh LB is loaded from the media port with Tween 20* and appropriate antibiotics. Flowing LB does not only permit a constant nutrition and exponentially growing cells for more than two days (that's the longest time we have tried), but also washes away newly generated cells to sustain a certain cell density, which is crucial for the determination of fluorescence strength. | ||
| - | + | </p> | |
| - | <b>Size</b> | + | <p><b>Size</b></p> |
<p> | <p> | ||
It has been stated before in the Mechanism part that most quorum sensing systems have a critical cell density requirement, so the trap size is designed neither too big nor too small to capture enough cells and allows for nutrients to permeate to every corner of the chamber. <br/><br/> | It has been stated before in the Mechanism part that most quorum sensing systems have a critical cell density requirement, so the trap size is designed neither too big nor too small to capture enough cells and allows for nutrients to permeate to every corner of the chamber. <br/><br/> | ||
You may also notice that the height of the chamber is extremely small, which is really demanding for the researcher's skills, but it's important for the observation. The oscillation could only be seen when the height of the chamber is in a particular range as our modeling predicted for many complicated reasons. For more details, please refer to the Modeling part. <br/><br/> | You may also notice that the height of the chamber is extremely small, which is really demanding for the researcher's skills, but it's important for the observation. The oscillation could only be seen when the height of the chamber is in a particular range as our modeling predicted for many complicated reasons. For more details, please refer to the Modeling part. <br/><br/> | ||
| - | </p> | + | </p><p> |
| - | <b>Material</b> | + | <b>Material</b></p> |
<p> | <p> | ||
Polydimethylsiloxane (PDMS), a transparent polymer material, is chosen to build the devise because of its excellent breathability, which is necessary for the permeation of H<sub>2</sub>O<sub>2</sub>. Once H<sub>2</sub>O<sub>2</sub> diffused outside cells, it can travel from colony to colony and synchronize them. <br/><br/> | Polydimethylsiloxane (PDMS), a transparent polymer material, is chosen to build the devise because of its excellent breathability, which is necessary for the permeation of H<sub>2</sub>O<sub>2</sub>. Once H<sub>2</sub>O<sub>2</sub> diffused outside cells, it can travel from colony to colony and synchronize them. <br/><br/> | ||
Our device is not only environmental friendly, but also a reusable one. After the flushing of a lysis buffer, most cells can be washed away, even those attached to walls. Then the device will be washed for three times by sterilized water before reuse.<br/><br/> | Our device is not only environmental friendly, but also a reusable one. After the flushing of a lysis buffer, most cells can be washed away, even those attached to walls. Then the device will be washed for three times by sterilized water before reuse.<br/><br/> | ||
| - | </p> | + | </p> |
| - | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/4/40/Xmumm-Image010.jpg" width=500 height=370 class="border" alt="" /> </td> | |
| - | + | </tr> | |
| - | + | <tr><td>Fig. 3-4 Microfluidic array- "Passage Model"</td></table></br> | |
| - | < | + | |
| - | + | <p><b>Image Capturing & Data analysis</b> </p> | |
<p> | <p> | ||
| - | The green fluorescence of bacteria in chambers is captured every 5 minutes by an inverted microscope (Eclipse Ti, Nikon) with a standard CCD camera manually and usually last more than 7 hrs. (This is really a big challenge for our team and very physically demanding since this is the only available fluorescence we could find. However, we managed to finish this data collecting process anyway. Good jobs, guys!) <br/><br/> | + | The green fluorescence of bacteria in chambers is captured every 5 minutes by an inverted microscope (Eclipse Ti, Nikon) with a standard CCD camera manually and usually last more than 7 hrs. (This is really a big challenge for our team and very physically demanding since this is the only available fluorescence we could find. However, we managed to finish this data collecting process anyway. Good jobs, guys!) <br/><br/> |
Data are transferred into oscillation curves by ImageJ and software designed by our teammate for digital image batch processing. (More detail for our self-designed software, please refer to protocol.) <br/><br/> | Data are transferred into oscillation curves by ImageJ and software designed by our teammate for digital image batch processing. (More detail for our self-designed software, please refer to protocol.) <br/><br/> | ||
</p> | </p> | ||
| - | + | <table><tr><td><img src="https://static.igem.org/mediawiki/2013/3/3e/Fig3_5.jpg" width="200" height="360" class="border" alt="" /> </td><td>Fig. 3-4 Microfluidic array- "Passage Model"</td></table> | |
| - | + | </tr> | |
| - | + | </table></br> | |
| - | + | <div class="clear"></div> | |
| - | + | <p>Data are transferred into oscillation curves by ImageJ and software designed by our teammate for digital image batch processing. (More detail for our self-designed software, please refer to <b>Software</b>.</p> | |
| - | + | <p>Next, we are going to <b>Improvement (For a better Glee)</b> to see how our little friends do on stage.</p> | |
| - | + | </div><!-- .center-block-page --> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | <p> | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <!-- END CONTENT --> | |
</div><!-- #wrapper --> | </div><!-- #wrapper --> | ||
| - | <!-- END WRAPPER --> | + | <!-- END WRAPPER --> |
| - | |||
| - | + | <div id="wrapper2"> | |
| - | + | ||
| - | + | <div id="separator"> | |
| + | |||
| + | <div class="center-block"> | ||
<a name="Modeling"></a> | <a name="Modeling"></a> | ||
| - | + | <h3>Modeling</h3> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <!-- START CONTENT --> | |
| - | + | <div class="center-block-page clearfix"> | |
| - | + | ||
| - | + | <h4>Mathematical model</h4> | |
| - | + | ||
| - | + | <p> | |
| - | We modeled the oscillate gene circuit (without <i>ndh</i> gene) based on the paper "A synchronized quorum of genetic clocks"<sup>[12]</sup>. In that, the author characterized the oscillator by using the four delay differential equations (DDEs), which are the basic of our modeling. DDE can perfectly simulate our circuit due to its powerful function about solving the dynamic change in biological processes.<br/><br/></p> | + | We modeled the oscillate gene circuit (without <i>ndh</i> gene) based on the paper "A synchronized quorum of genetic clocks"<sup>[12]</sup>. In that, the author characterized the oscillator by using the four delay differential equations (DDEs), which are the basic of our modeling. DDE can perfectly simulate our circuit due to its powerful function about solving the dynamic change in biological processes.<br/><br/></p> |
| - | + | <div class="clear"></div> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/d/d2/Xmumd-image01.jpg" class="border alignleft" alt="" /> | |
<div class="clear"></div> | <div class="clear"></div> | ||
<p> | <p> | ||
| - | + | In the first two equations, <img src="https://static.igem.org/mediawiki/2013/2/25/XMU-China_m2.png" style="height:30px"/> item is a Hill function equaling to <img src="https://static.igem.org/mediawiki/2013/e/e8/XMU-China_m1.png" style="height: 50px"/>, which describes the delayed reactions about the complex processes in the cells (transcription, translation, maturation, etc.). This Hill function takes the history of the system into account, i.e. the concentration of AHL at the time it binds to LuxR to form the activation complex.<br/> | |
This model does not include an equation for LuxR assuming that it is constitutively produced at a constant level. All parameters are normalized for the convenience of analysis.</p> <br /> | This model does not include an equation for LuxR assuming that it is constitutively produced at a constant level. All parameters are normalized for the convenience of analysis.</p> <br /> | ||
<h4>Modeling in MATLAB</h4><br /> | <h4>Modeling in MATLAB</h4><br /> | ||
<p>We used MATLAB to help us solve the DDEs and draw the graphs. In addition, we developed a GUI user interface after finishing the main DDE code. The user interface makes people easier to modify and adjust the parameters without recoding the .m files and any MATLAB language knowledge. We called this software "Gene OS" which is short for Gene oscillate simulator.</p><br /> | <p>We used MATLAB to help us solve the DDEs and draw the graphs. In addition, we developed a GUI user interface after finishing the main DDE code. The user interface makes people easier to modify and adjust the parameters without recoding the .m files and any MATLAB language knowledge. We called this software "Gene OS" which is short for Gene oscillate simulator.</p><br /> | ||
| - | <a href="https://2013.igem.org/Team:XMU-China/Software" style="font-size:20px">See more in Software</a> | + | <a href="https://2013.igem.org/Team:XMU-China/Software" style="font-size:20px">See more in Software</a> |
| - | + | </div><!-- .center-block-page --> | |
| - | |||
| - | + | ||
| + | <!-- END CONTENT --> | ||
</div><!-- #wrapper --> | </div><!-- #wrapper --> | ||
| - | <!-- END WRAPPER --> | + | <!-- END WRAPPER --> |
<div id="wrapper"> | <div id="wrapper"> | ||
| - | + | <div id="separator"> | |
| - | + | ||
| - | + | <div class="center-block"> | |
<a name="Application"></a> | <a name="Application"></a> | ||
| - | + | <h3>Applications</h3> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <!-- START CONTENT --> | |
| - | + | <div class="center-block-page clearfix"> | |
| - | + | ||
| - | + | <p> | |
After travelling a long way of circuit construction, you may wonder what we are going to do with those sparkling bacteria. Of course much thought was given, too, to make our oscillation circuit both showy and substantial as a safety guard in daily life. Here we are going to show you the potential of those shinny soldiers as<b style="font-size:20px"> biosensor, cancer cell killer and gene sinusoidal signal generator.</b> | After travelling a long way of circuit construction, you may wonder what we are going to do with those sparkling bacteria. Of course much thought was given, too, to make our oscillation circuit both showy and substantial as a safety guard in daily life. Here we are going to show you the potential of those shinny soldiers as<b style="font-size:20px"> biosensor, cancer cell killer and gene sinusoidal signal generator.</b> | ||
| - | </p> | + | </p> |
| - | + | <div class="clear"></div> | |
| - | + | <div class="clear"></div> | |
| - | + | ||
| - | + | ||
| - | + | </div><!-- .center-block-page --> | |
| + | |||
| + | <!-- END CONTENT --> | ||
<!-- START CONTENT --> | <!-- START CONTENT --> | ||
| - | + | <div class="center-block-page clearfix"> | |
| - | + | ||
| - | + | <h4>Biosensor</h4> | |
| - | + | ||
| - | + | <p> | |
To date there appears a great number of biosensors that are used for detecting certain factors, such as small organic molecules or metal ions. And we've noticed that each year many iGEM teams relate their projects to biosensors. Biological detection has strength in its low detection limit, high sensitivity and bioaffinity when compared with chemical detection methods. We can use the same gene sensing circuit to detect various metal ions or other determinant via changing different promoters which responds to a specific target molecule.<br/><br/> | To date there appears a great number of biosensors that are used for detecting certain factors, such as small organic molecules or metal ions. And we've noticed that each year many iGEM teams relate their projects to biosensors. Biological detection has strength in its low detection limit, high sensitivity and bioaffinity when compared with chemical detection methods. We can use the same gene sensing circuit to detect various metal ions or other determinant via changing different promoters which responds to a specific target molecule.<br/><br/> | ||
| - | + | Compared with traditional single-cell biosensor, our oscillatory system biosensor has three advantages. <br/><br/> | |
1) Synchronized colonies will be more sensitive to concentration changes of the target molecule and the changes in fluorescence strength are easier to observe than single cell biosensors. <br/><br/> | 1) Synchronized colonies will be more sensitive to concentration changes of the target molecule and the changes in fluorescence strength are easier to observe than single cell biosensors. <br/><br/> | ||
2) Single cell biosensor usually works, or "switches on" when the concentration of the specific molecules which can be responded by the promoter reaches to the lowest value. Sometimes this kind of detection is limited when we want to narrow the range of detection. Because the oscillation will happen only when concentration of certain molecule is between two value, the maximum and the minimum, it can dwindle the detection range of the determinant. <br/><br/> | 2) Single cell biosensor usually works, or "switches on" when the concentration of the specific molecules which can be responded by the promoter reaches to the lowest value. Sometimes this kind of detection is limited when we want to narrow the range of detection. Because the oscillation will happen only when concentration of certain molecule is between two value, the maximum and the minimum, it can dwindle the detection range of the determinant. <br/><br/> | ||
3) Oscillatory system is more robust and stable when used as a biosensor. These advantages will promote oscillatory gene circuits a better use in biosensing.<br/><br/> | 3) Oscillatory system is more robust and stable when used as a biosensor. These advantages will promote oscillatory gene circuits a better use in biosensing.<br/><br/> | ||
| - | + | ||
| - | + | </p> | |
<b>1. Arsenic sensing array</b> | <b>1. Arsenic sensing array</b> | ||
<br/><br/> | <br/><br/> | ||
<p>Jeff Hasty from UCSD used the same kind of oscillate circuit to engineer two kinds of arsenic-sensing macroscopic biosensors. These circuits are rewired by a native arsenite-responsive promoter, which is repressed by ArsR in the absence of arsenite. Due to the different mechanism, the sensor outputs are not the same. Figure 1 shows two constructed sensing modules and Figure 2 compares the outputs of two sensors<sup>[1]</sup>.<br/><br/> | <p>Jeff Hasty from UCSD used the same kind of oscillate circuit to engineer two kinds of arsenic-sensing macroscopic biosensors. These circuits are rewired by a native arsenite-responsive promoter, which is repressed by ArsR in the absence of arsenite. Due to the different mechanism, the sensor outputs are not the same. Figure 1 shows two constructed sensing modules and Figure 2 compares the outputs of two sensors<sup>[1]</sup>.<br/><br/> | ||
| - | </p> | + | </p> |
<div class="img-border"> | <div class="img-border"> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2013/f/f8/Xmua-Image003.png" width =340px height=240px class="border alignleft" alt="" /> | |
| - | + | </div> | |
| - | + | <div class="clear"></div> | |
<p>Figure 1.Two different constructed sensing modules.<br/></p> | <p>Figure 1.Two different constructed sensing modules.<br/></p> | ||
<div class="clear"></div> | <div class="clear"></div> | ||
<div class="img-border"> | <div class="img-border"> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2013/9/97/Xmua-Image005.jpg" width =200px height=200px class="border alignleft" alt="" /> | |
| - | + | </div> <div class="img-border"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/f/f6/Xmu-Image006.jpg" width =200px height=200px class="border alignleft" alt="" /> | |
| - | + | </div> | |
| - | + | <div class="clear"></div> | |
| - | <p>Figure 2. Two different outputs of corresponding biosensors.<br/></p> | + | <p>Figure 2. Two different outputs of corresponding biosensors.<br/></p> |
| - | + | <div class="clear"></div> | |
<b>2. Cytotoxin sensor</b> | <b>2. Cytotoxin sensor</b> | ||
<br/><br/> | <br/><br/> | ||
| - | <p>Due to the accuracy and specific condition in the field of bio-medicine, the oscillatory biosensor needs to meet the requirement such as synchronization. For example, all cells expected to twinkle at the same time only when cytotoxin existed. In addition, the period or amplitude of the oscillation needs to indicate the concentration of the cytotoxin. Our circuit has the potential to accomplish this kind of job. By further exploration in the relationship between oscillatory parameters such as period and amplitude with other environmental factors such as concentration of | + | <p>Due to the accuracy and specific condition in the field of bio-medicine, the oscillatory biosensor needs to meet the requirement such as synchronization. For example, all cells expected to twinkle at the same time only when cytotoxin existed. In addition, the period or amplitude of the oscillation needs to indicate the concentration of the cytotoxin. Our circuit has the potential to accomplish this kind of job. By further exploration in the relationship between oscillatory parameters such as period and amplitude with other environmental factors such as concentration of antibiotic or cytotoxin, we may create a oscillatory biosensor which can sense different kinds of biological toxin.<br/><br/> |
</p> | </p> | ||
| - | + | </div><!-- .center-block-page --> | |
| - | + | <div class="center-block-page clearfix"> | |
| - | + | ||
| - | + | <h4>Cancer cell killer</h4> | |
| - | + | <b>1.Cancer cell stalking </b> | |
| - | + | <p> | |
Nowadays cancer becomes one of the greatest enemy of our human being. Our main methods used to diagnosis of cancer are based on tissue and organ level, such as MRI, CT, PET, etc. These are mainly physical diagnostic methods.<br/><br/> | Nowadays cancer becomes one of the greatest enemy of our human being. Our main methods used to diagnosis of cancer are based on tissue and organ level, such as MRI, CT, PET, etc. These are mainly physical diagnostic methods.<br/><br/> | ||
With the development of science and technology, we create various methods into molecule level concentrated on molecular recognition and signal transduction of tumor marker, such as AFP(alpha fetoprotein), CEA (carcino-embryonic antigen), PSA(Prostate Specific Antigen) and so on. Level of these special molecules in human bodies has become the main basis of diagnosis of cancer. These are mainly chemical diagnostic methods.<br/><br/> | With the development of science and technology, we create various methods into molecule level concentrated on molecular recognition and signal transduction of tumor marker, such as AFP(alpha fetoprotein), CEA (carcino-embryonic antigen), PSA(Prostate Specific Antigen) and so on. Level of these special molecules in human bodies has become the main basis of diagnosis of cancer. These are mainly chemical diagnostic methods.<br/><br/> | ||
So how about using synthetic biology to do something for diagnosis of cancer? Our project can be a potential biological method to assist diagnosis of cancer.<br/><br/> | So how about using synthetic biology to do something for diagnosis of cancer? Our project can be a potential biological method to assist diagnosis of cancer.<br/><br/> | ||
ROS is reactive oxygen species in cells, which has been proved to be associated with several neurological disorders including Alzheimer's disease (AD) and Parkinson's diseases<sup>[2]</sup><sup>[3]</sup><sup>[4]</sup>. Concentration of ROS indicates the level of cell metabolism. Normally the stronger cell' metabolism is, the more ROS it will produce. In the recent decade scientist found that the occurrence of cancer is closely related to the level of ROS. But it is not a simple positive correlation as we expect<sup>[5]</sup>.<br/><br/> | ROS is reactive oxygen species in cells, which has been proved to be associated with several neurological disorders including Alzheimer's disease (AD) and Parkinson's diseases<sup>[2]</sup><sup>[3]</sup><sup>[4]</sup>. Concentration of ROS indicates the level of cell metabolism. Normally the stronger cell' metabolism is, the more ROS it will produce. In the recent decade scientist found that the occurrence of cancer is closely related to the level of ROS. But it is not a simple positive correlation as we expect<sup>[5]</sup>.<br/><br/> | ||
| - | </p> | + | </p> |
| - | + | <div class="clear"></div> | |
| - | + | <div class="img-border"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/2/2c/Xmua-Image008.png" width =200px height=200px class="border alignleft" alt="" /> | |
| - | + | </div> | |
| - | + | <div class="clear"></div> | |
| - | <p>From this picture we can get to know it clearly. As we know, we use | + | <p>Figure 4. ROS Level</p> |
| + | <p>From this picture we can get to know it clearly. As we know, we use oxide stress to describe the level of ROS in our cell. There is a threshold of oxide stress which is the critical value between normal cell and cancer cell. As a normal cell, its oxide stress is lower than the critical value while the ROS production value is over the threshold. On the one hand, when the oxide stress in our internal environment increases, normal cells may mutate to cancer cells if the ROS value is higher than the threshold value. On the other hand, cancer cells may die because of the accumulation of ROS. So the key point is, when ROS value higher than the maximum, we can kill the cancer cell while ROS value lower than the threshold we may push normal cells to change into cancer cells. So the level of ROS is in a specific range which we can measure using chemical methods.<br/><br/> | ||
Think about a proper range of ROS's concentration. Is it familiar to you as we mentioned a similar problem when talking about advantages of oscillatory system as biosensors? H<sub>2</sub>O<sub>2</sub> is a kind of ROS. The oscillation will happen only when the concentration of is H<sub>2</sub>O<sub>2</sub> between the maximum and the minimum. By regulating our circuits to let it respond in the proper concentration range which matches the ROS range of cancer cells, we can use the oscillatory system as a biological cancer detector. <br/><br/></p> | Think about a proper range of ROS's concentration. Is it familiar to you as we mentioned a similar problem when talking about advantages of oscillatory system as biosensors? H<sub>2</sub>O<sub>2</sub> is a kind of ROS. The oscillation will happen only when the concentration of is H<sub>2</sub>O<sub>2</sub> between the maximum and the minimum. By regulating our circuits to let it respond in the proper concentration range which matches the ROS range of cancer cells, we can use the oscillatory system as a biological cancer detector. <br/><br/></p> | ||
<div class="img-border"> | <div class="img-border"> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2013/0/0c/Xmu-a-a2.jpg" width=400px height=300px class="border alignleft" alt="" /> | |
| - | + | </div> | |
<p> | <p> | ||
| - | + | In our experiments, we found that we can regulate flow rate and some parameters of microfluidic chip such as the depth of the well, the width of the channel and the concentration of H<sub>2</sub>O<sub>2</sub> to change the period and amplitude of the oscillation. So by changing parameters of microfluidic chip, we can get a proper concentration range of H<sub>2</sub>O<sub>2</sub> if the oscillation works. From then on, we can extract some cancer tissue to detect the ROS concentration in the microfluidic chips or get some suspected tissue on chips and diagnose whether it contains cancer cell.<br/><br/> | |
</p> | </p> | ||
| - | + | <div class="clear"></div> <p> | |
<b>2 Periodical Killer</b><br/><br/><i>2.1 Drug delivery</i><br/><br/> | <b>2 Periodical Killer</b><br/><br/><i>2.1 Drug delivery</i><br/><br/> | ||
Our circuit also has a great potential application in periodic drug delivery. It is amazing when we realize that the oscillate circuit can control the drug to release in a specific period and the concentration of the drug is related to the amplitude. It is well-known that nowadays drug-release is a very hot research field. Many efforts are focusing on how to modify or delivery drug using materials such as synthetic polymers, liposome or nano shell which can respond to differences between different intracellular environments. Our oscillatory gene circuits provide a more biofriendly way for drug release. Our future plan is to make some deep exploration in this field.<br/><br/> | Our circuit also has a great potential application in periodic drug delivery. It is amazing when we realize that the oscillate circuit can control the drug to release in a specific period and the concentration of the drug is related to the amplitude. It is well-known that nowadays drug-release is a very hot research field. Many efforts are focusing on how to modify or delivery drug using materials such as synthetic polymers, liposome or nano shell which can respond to differences between different intracellular environments. Our oscillatory gene circuits provide a more biofriendly way for drug release. Our future plan is to make some deep exploration in this field.<br/><br/> | ||
| Line 988: | Line 531: | ||
In the applications of controlling pathogens, quorum sensing system plays an important role in regulating growth and reputation of microorganism. So we could restrain quorum sensing system to control microorganism to make our life more healthy and nice. The most important part in the metabolism of pathogens controlled by quorum sensing system is the synthesis of toxin and the formation of membrane system. If we can restrain these two kinds of effect may we decrease some potential diseases resources and improve the effects of antitoxins. What's more, it is a particularly attractive point to prevent pathogens through restraining their quorum sensing system instead of killing them directly as it offers the prospect of combating effectively infections resistant to all existing drugs. As we all know, just treat diseases via using antibotics or antitoxin can help these pathogens improve their resistance to various drugs, which is very dangerous for us human being. To date, although we've found that different fungus form different signal molecules, the quorum sensing system almost takes part in cell morphological transformation and information communication. We think our oscillatory gene circuits provide a very useful and meaningful model to promote more and more research in this field<sup>[11]</sup>.<br/><br/> | In the applications of controlling pathogens, quorum sensing system plays an important role in regulating growth and reputation of microorganism. So we could restrain quorum sensing system to control microorganism to make our life more healthy and nice. The most important part in the metabolism of pathogens controlled by quorum sensing system is the synthesis of toxin and the formation of membrane system. If we can restrain these two kinds of effect may we decrease some potential diseases resources and improve the effects of antitoxins. What's more, it is a particularly attractive point to prevent pathogens through restraining their quorum sensing system instead of killing them directly as it offers the prospect of combating effectively infections resistant to all existing drugs. As we all know, just treat diseases via using antibotics or antitoxin can help these pathogens improve their resistance to various drugs, which is very dangerous for us human being. To date, although we've found that different fungus form different signal molecules, the quorum sensing system almost takes part in cell morphological transformation and information communication. We think our oscillatory gene circuits provide a very useful and meaningful model to promote more and more research in this field<sup>[11]</sup>.<br/><br/> | ||
</p> | </p> | ||
| - | + | </div><!-- .center-block-page --> | |
| - | + | <!-- END CONTENT --> | |
<!-- START CONTENT --> | <!-- START CONTENT --> | ||
| - | + | <div class="center-block-page clearfix"> | |
| - | + | ||
| - | + | <h4>Gene sinusoidal signal generator</h4> | |
| - | + | <p> | |
With the development of science and technology, computer had been here and there with larger RAM, faster CPU and smaller volume. All these changes owe to VLSI (Very Large Scale Integrated Circuites), which consists of different electronic component.<br/><br/> | With the development of science and technology, computer had been here and there with larger RAM, faster CPU and smaller volume. All these changes owe to VLSI (Very Large Scale Integrated Circuites), which consists of different electronic component.<br/><br/> | ||
Just image what if we can control cell as a biocomputer. Cellular computer is a device consists of different kinds genetically engineered bacteria, which can do the calculation, such as add and subtract like a normal computer. To finish such a device, scientists need various gene circuits to complete different operations. These circuits are just like electronic component working in an electronic computer.<br/><br/> | Just image what if we can control cell as a biocomputer. Cellular computer is a device consists of different kinds genetically engineered bacteria, which can do the calculation, such as add and subtract like a normal computer. To finish such a device, scientists need various gene circuits to complete different operations. These circuits are just like electronic component working in an electronic computer.<br/><br/> | ||
At the beginning of this century, biocomputer is still a crazy dream far more than a possible blueprint. But now scientists have had the technology to make this "impossible mission" comes true. In recent months, scientists from Stanford constructed a new "bio-crystal valve" by DNA and RNA, which can do the logical operations in the living cells<sup>[6]</sup>.<br/><br/> | At the beginning of this century, biocomputer is still a crazy dream far more than a possible blueprint. But now scientists have had the technology to make this "impossible mission" comes true. In recent months, scientists from Stanford constructed a new "bio-crystal valve" by DNA and RNA, which can do the logical operations in the living cells<sup>[6]</sup>.<br/><br/> | ||
| - | + | Two months later, scientists from MIT used less than 3 gene devices to do the digital operation in division, logarithmic and square root. Thus, constructing gene circuits as electronic devices have become possible. From their research, their cellular calculator which uses analog computation has more advantages in computing efficiency in part count, speed and energy consumption compared with digital computation<sup>[7]</sup>. | |
| - | + | </p> | |
| - | + | <div class="clear"></div> | |
| - | + | <div class="img-border"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/0/09/Xmua-Image010.png" class="border alignleft" alt="" /> | |
| - | + | </div><div class="img-border"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/8/84/Image012.png" height=180px class="border alignleft" alt="" /> | |
| - | + | </div> | |
| - | + | <div class="clear"></div> | |
| - | <p>Figure 1: Different kinds of log-domain analog computation.</p> | + | <p>Figure 1: Different kinds of log-domain analog computation.</p> |
| - | + | <div class="clear"></div> | |
<p>Due to the fact that our circuits can generate a steady oscillation, it is possible that using our circuits as a sinusoidal signal generator.<br/><br/> | <p>Due to the fact that our circuits can generate a steady oscillation, it is possible that using our circuits as a sinusoidal signal generator.<br/><br/> | ||
Signal generators, such as function generator (including sinusoidal signal generator), are electronic devices that generate repeating or non-repeating electronic signals. As an important part of basic circuit studies, they are generally used in designing, testing, troubleshooting, and repairing electronic or electroacoustic devices. <br/><br/> | Signal generators, such as function generator (including sinusoidal signal generator), are electronic devices that generate repeating or non-repeating electronic signals. As an important part of basic circuit studies, they are generally used in designing, testing, troubleshooting, and repairing electronic or electroacoustic devices. <br/><br/> | ||
| Line 1,016: | Line 559: | ||
<div class="clear"></div> | <div class="clear"></div> | ||
<div class="img-border"> | <div class="img-border"> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2013/0/05/Xmua-Image013.png"class="border alignleft" alt="" /> | |
| - | + | </div> | |
<div class="clear"></div> | <div class="clear"></div> | ||
<p>Figure 2.This picture shows a kind of electronic circuit generating the oscillation wave.</p> | <p>Figure 2.This picture shows a kind of electronic circuit generating the oscillation wave.</p> | ||
<div class="clear"></div> | <div class="clear"></div> | ||
<div class="img-border"> | <div class="img-border"> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2013/2/22/Xmua-Image015.png"class="border alignleft" alt="" /> | |
| - | + | </div> | |
<div class="clear"></div> | <div class="clear"></div> | ||
<p>Figure 3.This graph shows the generated oscillation wave.</p> | <p>Figure 3.This graph shows the generated oscillation wave.</p> | ||
| - | + | </div><!-- .center-block-page --> | |
| - | + | <!-- END CONTENT --> | |
<!-- START CONTENT --> | <!-- START CONTENT --> | ||
| - | + | <div class="center-block-page clearfix"> | |
| - | + | ||
| - | + | <h4>Future Version Trilogy</h4> | |
| - | + | <p> | |
We blueprint our next step developing oscillatory gene circuits in following steps:<br/><br/> | We blueprint our next step developing oscillatory gene circuits in following steps:<br/><br/> | ||
<b>Step One<br/><br/> | <b>Step One<br/><br/> | ||
| Line 1,042: | Line 585: | ||
Confirm most suitable microfluidic parameters for oscillation;<br/> | Confirm most suitable microfluidic parameters for oscillation;<br/> | ||
Optimize our model<br/> | Optimize our model<br/> | ||
| - | </p> | + | </p> |
| - | + | <div class="clear"></div> | |
<p><b>Step Two<br/><br/> | <p><b>Step Two<br/><br/> | ||
We will make our oscillation a controllable one by integrating the catalase gene into the circuit.<br/><br/></b> | We will make our oscillation a controllable one by integrating the catalase gene into the circuit.<br/><br/></b> | ||
Catalase is a common enzyme found in nearly all living organisms exposed to oxygen. It catalyzes the decomposition of hydrogen peroxide to water and oxygen. It is a very important enzyme in protecting the cell from oxidative damage by reactive oxygen species (ROS). Since our lab is studying <i>Shewanella oneidensis</i>, and its catalase protein has showed outstanding performance in catalyzing H<sub>2</sub>O<sub>2</sub>. So we thought we could integrate the catalase gene into our circuit and under an artificial inducible promoter, by which we can decide when to start this catalase gene and stop the oscillation.<br/><br/> | Catalase is a common enzyme found in nearly all living organisms exposed to oxygen. It catalyzes the decomposition of hydrogen peroxide to water and oxygen. It is a very important enzyme in protecting the cell from oxidative damage by reactive oxygen species (ROS). Since our lab is studying <i>Shewanella oneidensis</i>, and its catalase protein has showed outstanding performance in catalyzing H<sub>2</sub>O<sub>2</sub>. So we thought we could integrate the catalase gene into our circuit and under an artificial inducible promoter, by which we can decide when to start this catalase gene and stop the oscillation.<br/><br/> | ||
| - | + | </p> | |
<p> | <p> | ||
<b>Step Three<br/><br/> | <b>Step Three<br/><br/> | ||
| Line 1,053: | Line 596: | ||
Make our circuit function as biosensor, cancer cell killer and gene sinusoidal signal generator.<br/><br/> | Make our circuit function as biosensor, cancer cell killer and gene sinusoidal signal generator.<br/><br/> | ||
</p> | </p> | ||
| - | + | <div class="clear"></div> | |
| + | <div class="center-block-page clearfix"> | ||
| + | <b><p>Reference</p></b> | ||
| + | <p> | ||
| + | 1. Prindle, A., et al., A sensing array of radically coupled genetic 'biopixels'. <i>Nature</i> 481, 39-44 (2012).<br/> | ||
| + | 2. Gaggelli, E. Kozlowski, H. Valensin, D. Valensin & G. Copper ,Homeostasis and Neurodegenerative Disorders (Alzheimer's,Prion, and Parkinson's Diseases and Amyotrophic Lateral Sclerosis). <i>Chem. Rev.</i> 106, 1995–2044 (2006).<br/> | ||
| + | 3. Scott, L. E. Orvig, C. Medicinal Inorganic Chemistry Approaches to Passivation and Removal of Aberrant Metal Ions in Disease. <i>Chem. Rev.</i> 109, 4885–4910 (2009).<br/> | ||
| + | 4. Zheng, Z. Q. White, C. Lee, J. Peterson, T. S. Bush, A. I. Sun, G. Y. Weisman, G. A. & Petris, M. J. Altered Microglial Copper Homeostasis in a Mouse Model of Alzheimer's Disease. <i>J. Neurochem.</i> 114, 1630–1638 (2010).<br/> | ||
| + | 5. Dunyaporn T., Jerome A. & Peng H., Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? <i>Nature Reviews Drug Discovery</i> 8, 579-591 (2009).<br/> | ||
| + | 6. Bonnet J, Yin P, Ortiz ME, Subsoontorn P & Endy D, Amplifying Genetic Logic Gates. <i>Science</i> 340, 599-603 ,2013.<br/> | ||
| + | 7. Ramiz D., Timothy K. Lu et al. Synthetic analog computation in living cells. <i>Nature</i> 497, 619-+ (2013).<br/> | ||
| + | 8. Guang bi Li et al, Electronic Ciircuit Model Construction of A Synthetic Gene Network. <i>Journal of Tianjin University of Science & Technology</i> 26, 22-25 (2011).<br/> | ||
| + | 9. Hasty, J., Dolnik, M., Rottschafer, V. & Collins, J. J. Synthetic gene network for entraining and amplifying cellular oscillations. <i>Physical Review Letters</i> 88, 148101, (2002).<br/> | ||
| + | 10. Elowitz, M. B. & Leibler, S. A. Synthetic oscillatory network of transcriptional regulators. <i>Nature</i> 403, 335 (2000).<br/> | ||
| + | 11. Yan C., Quorum--Sensing System in Microbes and Its Application, School of Resources and Environment, Yuxi Normal University,Yuxi,Yunnan 653100.<br /> | ||
| + | 12. Danino, T., Mondragon-Palomino, O., Tsimring, L. & Hasty, J., A synchronized quorum of genetic clocks. <i>Nature</i> 463, 326-330 (2010).<br/> | ||
| + | </p> | ||
| - | + | <div class="clear"></div> | |
| - | + | </div><!-- .center-block-page --> | |
| - | </div><!-- | + | |
| - | <!-- | + | </div><!-- .center-block-page --> |
| - | <!-- | + | <!-- END CONTENT --> |
| - | <div | + | </div><!-- #wrapper --> |
| - | + | <!-- END WRAPPER --> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | < | + | <!-- START WRAPPER --> |
| - | < | + | <div id="wrapper1"> |
| - | < | + | <div id="separator"> |
| - | + | ||
| - | < | + | <div class="center-block"> |
| - | + | ||
| - | < | + | <h3>Parts</h3> |
| - | < | + | </div> |
| + | </div> | ||
| - | <div class=" | + | <!-- START CONTENT --> |
| + | <div class="center-block-page clearfix"> | ||
| - | < | + | <h4>Table 1. The Bio-bricks used in the study</h4> |
| - | + | ||
| - | < | + | <table class=MsoTableGrid border=1 cellspacing=0 cellpadding=0 width=950 |
| + | > | ||
| + | <tr> | ||
| + | <td width=105 rowspan=2 > | ||
| + | <p ><b >Part<o:p></o:p></span></b></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=102 rowspan=2> | ||
| + | <p ><span | ||
| + | ><b>Backbone<o:p></o:p></span></b></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=106 rowspan=2 > | ||
| + | <p ><span | ||
| + | ><b>Type<o:p></o:p></span></b></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=99 rowspan=2 > | ||
| + | <p ><span | ||
| + | ><b>Location<o:p></o:p></span></b></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=161 colspan=2> | ||
| + | <p ><span | ||
| + | ><b>Size | ||
| + | (bp)<o:p></o:p></span></b></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=377 rowspan=2> | ||
| + | <p ><span | ||
| + | ><b>Description<o:p></o:p></span></b></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | </tr> | ||
| + | <tr style='mso-yfti-irow:1;height:7.85pt'> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><b>part<o:p></o:p></span></b></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><b>backbone<o:p></o:p></span></b></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | </tr> | ||
| + | <tr style='mso-yfti-irow:2'> | ||
| + | <td width=105> | ||
| + | <p ><span | ||
| + | ><span >BBa_K546000<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=102> | ||
| + | <p ><span | ||
| + | >pSB1C3<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=106 > | ||
| + | <p ><span | ||
| + | >Signaling</span></span><span ><span | ||
| + | lang=EN-US style='font-size:11.0pt;mso-bidi-font-size:12.0pt;font-family: | ||
| + | "Times New Roman","serif"'><o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=99> | ||
| + | <p ><span | ||
| + | ><span lang=EN-US >2013-P1-12D<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><span >1964<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><span >2070<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=377 > | ||
| + | <p ><span | ||
| + | ><span >Lux pL | ||
| + | controlled LuxR with lux pR autoinducing LuxI (lva tag)- AHL.<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | </tr> | ||
| + | <tr style='mso-yfti-irow:3'> | ||
| + | <td width=105> | ||
| + | <p ><span | ||
| + | ><span >BBa_I763020<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=102> | ||
| + | <p ><span style='mso-bookmark: | ||
| + | _Hlk367271772'><span>pSB1C3</span></span><span | ||
| + | ><span lang=FR ><o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=106 > | ||
| + | <p ><span style='mso-bookmark: | ||
| + | _Hlk367271772'><span >Intermediate<o:p></o:p></span></span></p> | ||
| + | <p><span style='mso-bookmark: | ||
| + | _Hlk367271772'><span >RBS-GFP-TT<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=99> | ||
| + | <p ><span | ||
| + | ><span lang=EN-US >2013-P3-11H<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><span >914<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><span >2070<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=377 > | ||
| + | <p ><span | ||
| + | ><span >RBS-GFP | ||
| + | (+LVA Tag)-Terminators<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | </tr> | ||
| + | <tr style='mso-yfti-irow:4'> | ||
| + | <td width=105> | ||
| + | <p ><span | ||
| + | ><span >BBa_F2621<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=102> | ||
| + | <p ><span | ||
| + | >pSB1A2<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=106 > | ||
| + | <p ><span | ||
| + | >Signaling</span></span><span ><span | ||
| + | lang=EN-US style='font-size:11.0pt;mso-bidi-font-size:12.0pt;font-family: | ||
| + | "Times New Roman","serif"'><o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=99> | ||
| + | <p ><span | ||
| + | ><span lang=EN-US >2013-P2-21F<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><span >1158<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><span >2079<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=377 > | ||
| + | <p ><span | ||
| + | ><span >3OC6HSL | ||
| + | Receiver Device<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | </tr> | ||
| + | <tr style='mso-yfti-irow:5;height:39.75pt'> | ||
| + | <td width=105 > | ||
| + | <p ><span | ||
| + | ><span >BBa_K546001<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=102 > | ||
| + | <p ><span | ||
| + | ><span >pSB1C3</span></span><span ><span | ||
| + | lang=EN-US style='font-size:11.0pt;mso-bidi-font-size:12.0pt;font-family: | ||
| + | "Times New Roman","serif"'><o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=106 > | ||
| + | <p ><span | ||
| + | ><span >Device<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=99> | ||
| + | <p ><span | ||
| + | ><span lang=EN-US >2013-P1-12F<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81> | ||
| + | <p ><span | ||
| + | ><span >2135<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81> | ||
| + | <p ><span | ||
| + | ><span >2070<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=377 > | ||
| + | <p ><span | ||
| + | ><span >AIIA<o:p></o:p></span></span></p> | ||
| + | <p ><span | ||
| + | ><span >AHL Reporter | ||
| + | and Quencher<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width=105> | ||
| + | <p ><span >BBa_J04450<o:p></o:p></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=102 > | ||
| + | <p ><span | ||
| + | ><span >pSB4K5<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=106 > | ||
| + | <p ><span | ||
| + | ><span >Reporter<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=99 > | ||
| + | <p ><span | ||
| + | ><span | ||
| + | ><span>2013-P5-5G<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81> | ||
| + | <p ><span | ||
| + | ><span >1069<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81> | ||
| + | <p ><span | ||
| + | ><span >3419<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=377 > | ||
| + | <p ><span | ||
| + | ><span >RFP Coding | ||
| + | Device<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | </tr> | ||
| + | <tr style='mso-yfti-irow:7;height:13.15pt'> | ||
| + | <td width=105 > | ||
| + | <p ><span | ||
| + | ><span >BBa_J04450</span></span><span | ||
| + | ><span ><o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=102 > | ||
| + | <p ><span | ||
| + | ><span >pSB3T5<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=106 > | ||
| + | <p ><span | ||
| + | ><span >Reporter<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=99 > | ||
| + | <p ><span | ||
| + | ><span >2013-P5-7C<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><span >1069<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=81 > | ||
| + | <p ><span | ||
| + | ><span lang=FR >3241<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | <td width=377 > | ||
| + | <p ><span | ||
| + | ><span lang=FR >RFP Coding Device<o:p></o:p></span></span></p> | ||
| + | </td> | ||
| + | <span ></span> | ||
| + | </tr> | ||
| + | <tr '> | ||
| + | <td width=105 > | ||
| + | <p ><span><span | ||
| + | >BBa_J23022</span><o:p></o:p></span></p> | ||
| + | </td> | ||
| + | <td width=102 > | ||
| + | <p ><span >BBa_J23006<o:p></o:p></span></p> | ||
| + | </td> | ||
| + | <td width=106 > | ||
| + | <p ><span >Composite<o:p></o:p></span></p> | ||
| + | </td> | ||
| + | <td width=99 > | ||
| + | <p ><span >2013-P5-3H<o:p></o:p></span></p> | ||
| + | </td> | ||
| + | <td width=81 > | ||
| + | <p ><span >234<o:p></o:p></span></p> | ||
| + | </td> | ||
| + | <td width=81 > | ||
| + | <p ><span lang=FR | ||
| + | >2356<o:p></o:p></span></p> | ||
| + | </td> | ||
| + | <td width=377 > | ||
| + | <p ><span lang=FR | ||
| + | >[key3d][TT]<o:p></o:p></span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| - | < | + | </br></br> |
| + | <table width="100%" border="1" cellspacing=0> | ||
| + | <b><p> | ||
| + | Favorite XMU-China 2013 iGEM Team Parts | ||
| + | <p></b> | ||
| + | <tr> | ||
| + | <th scope="col">W/N</th> | ||
| + | <th scope="col">Name</th> | ||
| + | <th scope="col">Type</th> | ||
| + | <th scope="col">Description</th> | ||
| + | <th scope="col">Designer</th> | ||
| + | <th scope="col">Length</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>W</td> | ||
| + | <td>BBa_K1036000</td> | ||
| + | <td>Composite</td> | ||
| + | <td>lux pL controlled luxR with lux pR controlled ndh (LVA-tag) coding for NADH dehydrogenase II</td> | ||
| + | <td>Shengquan Zeng</br> Zhaopeng Cheng</td> | ||
| + | <td>2687</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>W</td> | ||
| + | <td>BBa_K1036001</td> | ||
| + | <td>Coding</td> | ||
| + | <td>the ndh gene coding for respiratory NADH dehydrogenase II</td> | ||
| + | <td>Shengquan Zeng</br> Zhaopeng Cheng</td> | ||
| + | <td>1366</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>W</td> | ||
| + | <td>BBa_K1036003</td> | ||
| + | <td>Composite</td> | ||
| + | <td>lux pL controlled luxR with lux pR controlled gfp (LVA-tag)</td> | ||
| + | <td>Xin Guo</br>Xi Xi</td> | ||
| + | <td>4052</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>W</td> | ||
| + | <td>BBa_K1036006</td> | ||
| + | <td>Composite</td> | ||
| + | <td>lux pL controlled luxR with lux pR controlled sfgfp (with LVA-tag)</td> | ||
| + | <td>Zhaopeng Cheng</br>Xiyu Wu</td> | ||
| + | <td>4046</td> | ||
| + | </tr> | ||
| + | </table> | ||
| - | < | + | </br></br> |
| + | <table width="100%" border="1" cellspacing=0> | ||
| + | <b><p> | ||
| + | XMU-China 2013 iGEM Team Parts Sandbox | ||
| + | <p></b> | ||
| + | <tr> | ||
| + | <th scope="col">W/N</th> | ||
| + | <th scope="col">Name</th> | ||
| + | <th scope="col">Type</th> | ||
| + | <th scope="col">Description</th> | ||
| + | <th scope="col">Designer</th> | ||
| + | <th scope="col">Length</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>W</td> | ||
| + | <td>BBa_K1036002</td> | ||
| + | <td>Composite</td> | ||
| + | <td>gfp (with LVA-tag) under plux R control</td> | ||
| + | <td>Xin Guo</br> Xi Xi</td> | ||
| + | <td>2866</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>W</td> | ||
| + | <td>BBa_K1036004</td> | ||
| + | <td>Reporter</td> | ||
| + | <td>sfgfp with LVA-tag</td> | ||
| + | <td>Zhaopeng Cheng</br>Xiyu Wu</td> | ||
| + | <td>753</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>W</td> | ||
| + | <td>BBa_K1036005</td> | ||
| + | <td>Composite</td> | ||
| + | <td>sfgfp (with LVA-tag) under plux R control</td> | ||
| + | <td>Zhaopeng Cheng</br>Xiyu Wu</td> | ||
| + | <td>2880</td> | ||
| + | </tr> | ||
| - | < | + | </table> |
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | </div><!-- .center-block-page --> | |
| + | |||
| + | <!-- END CONTENT --> | ||
</div><!-- #wrapper --> | </div><!-- #wrapper --> | ||
| Line 1,682: | Line 1,056: | ||
<!-- START WRAPPER --> | <!-- START WRAPPER --> | ||
| - | < | + | <!-- #wrapper --> |
| - | < | + | <!-- END WRAPPER --> |
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<!-- START FOOTER --> | <!-- START FOOTER --> | ||
| - | + | <div class="clear"></div> | |
| - | + | <div id="home_s4"><center> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/c/cb/Xmusoftware_sponsor1.png" width="80px" height="80px"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/1/18/Xmusoftware_sponsor2.png" width="80px" height="80px"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/6/6f/Xmu-Bg_header_mwlogo_notag.jpg" height="60px"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/e/ed/Xmu-evernote.jpg" height="60px"> | |
| - | + | </center> | |
</div> | </div> | ||
<div id="footer-menu"> | <div id="footer-menu"> | ||
| - | + | <div class="center-block clearfix"> | |
| - | + | <ul id="second-menu"> | |
| - | + | <li><a href="https://2013.igem.org/Team:XMU-China">Home</a></li> | |
| - | + | <li><a href="https://2013.igem.org/Team:XMU-China/Team1">Team</a></li> | |
| - | + | <li><a href="https://2013.igem.org/Team:XMU-China/Project">Project</a></li> | |
| - | + | <li><a href="https://2013.igem.org/Team:XMU-China/Notebook">Notebook</a></li> | |
| - | + | <li><a href="https://2013.igem.org/Team:XMU-China/Software">Software</a></li> | |
| - | + | <li><a href="https://2013.igem.org/Team:XMU-China/Human Practice">Human Practice</a></li> | |
| - | + | <li><a href="https://2013.igem.org/Team:XMU-China/Safety">Safety</a></li> | |
| - | + | </ul> | |
| - | + | <div class="copyright">© Copyright © 2013.XMU-China iGEM All rights reserved.</div> | |
| - | + | </div><!-- .center-block --> | |
</div><!-- #footer-menu --> | </div><!-- #footer-menu --> | ||
| - | <!-- END FOOTER MENU | + | <!-- END FOOTER MENU --> |
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 01:54, 26 October 2013
Forming the Glee
To be a good glee, first it needs a good song. The most intriguing song on earth is called OSCILLATION. If you look about, you will find it ubiquitous and extremely useful. After we taught each member this song, we need to help them sing it together, so oscillation became SYNCHRONIZED OSCILLATION. Thus how we form a glee!
Oscillation: The Magic Song of World
Once upon the time, there was a magic song called oscillation, it permeated every corner of the world. It regulated nearly everything on the earth. The interaction of moon and ocean generated the magnificent tide oscillation, and the biological process that displays a 24-hour oscillation inside human body.
 |

|
| Fig. 1-1 The Tides depend on several factors, including where the sun and moon are relative to the Earth | Fig. 1-2 The circadian clock inside human body |
Besides, oscillation also plays an important part in signal transition. Take lasers for example, which are known for their intensity and can be focused to a tight spot over long distance. These characteristics all owe to their spatial coherent in the frequency of the light source.
 |
| Fig. 1-3 Helium-Neon laser demonstration at the Kastler-Brossel Laboratory at Paris VI: Pierre et Marie |
Oscillation in Bacteria
Synthetic biologists have done a lot of work to teach bacteria to sing this oscillation song by building artificial genetic circuit inside them, since oscillations can lead to fantastic applications and benefit our everyday life. Besides regulation and signal transmission, oscillations in living organisms can also react to its growing environment, which probably will activate the trigger of oscillation inside cells and generate oscillation signals. From collected signals we can tell how an environment factor influences the cells behavior, and in turn the environment factor can be indicated by the signals. Furthermore, if we interpret these kinds of oscillation signals like 1010 in binary system, then we could even make an electronic environmental factor detector, using signals from environment as an input, and numbers on computer screen as output (Fig. 1-4).
 |
| Fig. 1-4 How to make a signal converter |
Here is the progress of artificial synthetic biological oscillations have made.
In the first generation of oscillation, three transcriptional repressor systems that are not part of any natural biological clock are used to build an oscillating network, termed the repressilator, in Escherichia coli (Fig. 1-5). The network periodically induces the synthesis of green fluorescent protein as a readout of its state in individual cells. However, plasmids can hardly pass from generation to generation and this artificial clock is not robust enough and always displays noisy behavior.
 |
| Fig. 1-5 The first generation oscillator, known as repressor |
Scientists constructed the second generation oscillator rapidly. This improved oscillator is fast, robust and persistent; with tunable oscillatory periods as fast as 13 min (Fig.1-6). The oscillator is designed using a previously modeled network architecture comprising linked positive and negative feedback loops. But this circuit could only monitor oscillations in individual cells through multiple cycles, which means the song is still limited to single individual.
 |
| Fig. 1-6 2nd generation of oscillation consists of positive and negative feedback loops |
Thinking that singing alone is lonely, scientists introduce quorum sensing to connect isolated cells and let them sing together, and technically it's called synchronized oscillation. The synchronization effect of quorum sensing, however, is limited to over tens of micrometer (Fig.1-7). If the synchronized colony could be enlarged, then the oscillation signal will be much stronger and steadier. Longer ranged synchronized oscillation is realized by a gas-phase redox signal H2O2 to the third generation circuit, which could achieve the communication among colonies at over millimeter scales and became the fourth generation. Oh yeah, finally our bacteria can sing the same oscillation at the same time, so they decided to form their own glee!
 |
| Fig.1-7 3rd generation involved in quorum sensing |
That's how our little E. coli friend formed their glee, a long but cheerful story. Thanks to hardworking synthetic biologists. Let's go and see how this fourth generation oscillation glee functions inside in THREE PLASMIDS!
Reference
1. http://en.wikipedia.org/wiki/Oscillation
2. Elowitz M. B. & Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335-338 (2000)
3. Stricker J. et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516-519 (2008)
4. Danino T., Mondrago´n-Palomino O., Tsimring L. & Hasty J. A synchronized quorum of genetic clocks. Nature 463, 326-330 (2010)
Three Parts in Circuit Designation
In a synchronized oscillatory system, three important parts should be included: the oscillator, which is the biochemical machinery that generate the oscillatory output; the coupling pathway that ensure the connection among cells; and output pathway, which is also known as a reporter that reflect the state of the oscillator to downstream targets (Fig. 2-1).
 |

|
| Fig. 2-1 Circuit working mechanism |
Oscillator
Quorum sensing (QS) is a cell-to-cell signaling mechanism that refers to the ability of bacteria to respond to chemical hormone-like molecules called autoinducers. When an autoinducer reaches a critical threshold, the bacteria detect and respond to this signal by altering their gene expression. In our circuit, QS (from Vibrio fischeri) is installed as a positive feedback while aiiA (from Bacillus Thurigensis) acts as a negative one to compose an oscillator together. In the quorum sensing part, the luxI gene is at low expression level and produces LuxI protein that synthesize a kind of acyl-homoserine lactone (AHL), which is a small molecule that can diffuse across the cell membrane and mediate intercellular coupling when it reaches the threshold as enough biomass accumulated. AHL will bind intracellular protein LuxR, which is also consecutively produced by luxR gene. The LuxR-AHL complex can activate the luxI promoter, and the positive feedback loop is built. At the same time, the aiiA gene, which is under control of the same promoter is expressed and produce a lactonase enzyme known as aiiA that hydrolyzes the lactone ring of AHL. (Fig. 2-1) In this system, the activator enhances the expression of both activator and repressor, which shares the common motif of many synthetic oscillators.
Coupling
The communication range of quorum sensing, however, is limited by the diffusion rate of AHL, could only reach cells over tens of micrometers. So, we introduced the coding gene for NADH dehydrogenase II (ndh) and put it under the control of the same luxIpromoter, which means ndh also has a periodical expression in accordance with the oscillator. NDH-2 is a membrane-bound respiratory enzyme that generates low level H2O2 and superoxide (O2-) and H2O2 will permeate to neighboring colonies. Periodic production of H2O2 changes the redox state of a cell immediately and interacts with the synthetic circuit through the native aerobic response control systems, including ArcAB, which has a binding site in the lux promoter region. Before the oxidizing condition is triggered, ArcAB is partially expressed, so lux is partially repressed. When the concentration of H2O2 is increasing, ArcAB is gradually inactivated, and the repression of lux is relieved. With the periodically produced vapor phase H2O2 that can diffuse quickly among colonies, the oscillation is not only strengthened but also expanded to millimeter scales(Fig. 2-2).
 |
| Fig. 2-2 H2O2 functions as an activator to the quorum sensing promoter |
When compared with the strong but short ranged coupling by QS, H2O2 might be weaker but long ranged because its disperse characteristic. These two levels of communication between cells formed the basic oscillatory system inside our host.
Reporter
A good reporter in an oscillatory circuit should be steadily expressed and can be easily detected by regular equipment in laboratory, so broadly used reporter green fluorescent protein (GFP) came into our mind.
First, we built regular gfp gene into our circuit and it worked well. Its fluorescence can be observed by fluorescence microscopy under 1s of exposure time on the microfluidic array 1(Fig. 2-3).
 |
| Fig. 2-3 GFP under fluorescence microscopy on a microfluidic array |
However, there is a newly found GFP variant Superfolder GFP, which has shown outstanding performance in many ways. It has two advantages in observing:
1) Fold 3.5 to 4 times faster than regular GFP;
2) Yield up to four times more total Fluorescence than regular GFP.
The greater the fluorescence strength is, the shorter exposure time will be needed, thus can decrease the photobleaching (GFP will be eventually destroyed by the light exposure that tries to stimulate it into fluorescing) that will lead into lapses in data processing.
Besides, we also want to make a comparison between two different GFPs' performances in our oscillatory circuit. For example, peaks, troughs and periods in oscillatory curves. The following picture shows the difference between GFP and sfGFP both on colE1 backbone.(Fig. 2-4)
 |
| Fig.2-4 Difference between GFP and sfGFP both on colE1 backbone (P. S. Thanks the Peking iGEM team 2013 for providing us the sfGFP part.) |
Please come with us to see how we built our magic circuit in E. coli in Construction!
Overview
So we designed three plasmids to function as the three important parts in the oscillation circuit, and all of them are in the charge of the same quorum sensing promoter to make sure that all target genes share the same oscillatory period (Table 2-1).
| No. | Plasmid | ReplicationOrigin | CopyNumber | Resistance | Size(bp) | |
|---|---|---|---|---|---|---|
| Insert | Backbone | |||||
| A1 | pSB1C3-gfp-luxI | pSB1C3 | high(100~300) | Chloromycetin(Cm) | 4052 | 2070) |
| A2 | pSB1C3-sfgfp-luxI | pSB1C3 | high(100~300) | Chloromycetin(Cm) | 4046 | 2070) |
| A3 | p3H-GFP-luxI | p3H | Middle(18~22) | Ampicillin (Amp) | 4052 | / |
| A4 | p3H-sfGFP-luxI | p3H | Middle(18~22) | Ampicillin (Amp) | 4046 | / |
| B | pSB3T5-aiiA | p15A | Middle(10~12) | Tetracycline (Tet) | 2135 | 2837 |
| C | pSB4K5-ndh | pSC101 | Low(5) | Kanamycin (Kan) | 2658 | 3004 |
In a synchronized oscillation circuit, each A, B and C plasmid is necessary. The plasmid A expresses GFP breporter and LuxI proteins, which is the positive feedback in oscillator; the plasmid B expresses protein aiiA to degrade AHL and acts as the negative feedback in oscillator; the plasmid C expresses NAD-2 to generate H2O2 to communicate between colonies, is the coupling part in circuit (Fig 2-5).
 |
| Fig. 2-5 Plasmids A, B and C in oscillation circuit |
You may have noticed that these three plasmids have different resistant genes and replication origins on their backbone. Different resistant genes are used mainly for selection. And since different replication origin means a different copy number of target genes in this plasmid. Using different replication origins is to produce different target proteins in an appropriate ratio, and only in this way can oscillation be observed. Just like a glee needs Soprano, Tenor and Bass to cooperate to finish a song. Besides, three artificial plasmids with same copy numbers always have a competitive relation in one cell and cannot be well expressed.
Plasmid A has two different backbones with high and middle copy numbers respectively. We built them to see the copy numbers' effect on oscillation. Results can be seen in Improvement (For a better Glee).
All of the plasmids we constructed are confirmed by agarose gel electrophoresis, and result can be seen in Parts, where you can also get the link to parts we submitted.
After the onerous construction work, our glee is ready to perform the synchronized oscillation song, so we have to find them a stage. Let's go to see the beautiful opera in Microfluidics (Stages).
Reference
1. Prindle, A., et al., A sensing array of radically coupled genetic 'biopixels'. Nature 481, 39-44, 2012;
2. Pedelacq, J. et al., Engineering and characterization of a superfolder green fluorescent protein. Nature 24, 79-88, 2006;
3. Waters C. M. & Bassler B. L. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol 21, 319-46, 2005.
Microfluidics (Opera Houses)
A Microfluidic array to cells is like an opera house to a glee that provides a cell culture environment and observation stage. To accompany the observation of microfluidic we used the fluorescence microscope, just like mass media to photograph the oscillation. We call them M & M. What a lovely couple.
Why M&M?
If we want to capture gene expression dynamics and distributions reported by fluorescence, we need to create a chemostat that provide a near-constant environment for the growth of bacteria. The near-constant environment mainly includes two aspects: continuous fresh media to ensure the optimum growth state and constant small populations of bacteria to ensure single-cell resolution in observation. Steady growth state is needed because the oscillation curve is expected to be a near simple harmonic one. Besides, bacteria tend to form biofilm which will lead to the overlapping of cells and create a false impression that the fluorescence increases, and that's why we have to prevent this to maintain a constant bacteria population with single-cell resolution.
What we mentioned above might have covered all aspects we need to consider for communication in a single colony, but if we still want to observe the coupling between colonies we have to create an array that contains many colonies and allows H2O2 to pass through freely.
Among many different techniques, microfluidic array seems to be our best choice with following advantages:
1) Precisely control of flowing rate and cell populations when equipped with syringe pumps;
2) Cells behavior including fluorescence strength can be captured by high resolution microscopy;
3) Trap numbers and dimensions can be adapted to different experimental need;
4) Productive: fewer resources and less time are required when compared with macroscopic cultures.
Design of Microfluidic Array
1.Pretests in "Swimming Pool"
Before our experiments on the microfluidic array we described before, a series of pretests were done on a chip whose structure looks like many swimming pool on the ground (Fig. 3-1).
 |
| Fig. 3-1 A "Swimming pool" chip in black field; those white squares (100μm×100μm×100μm) are chambers for bacteria |
From the sketch (Fig. 3-2) we can see that the chamber is like a swimming pool where bacteria swim happily in the fresh LB that running from up above, and newly generated cells would be washed away.
 |
| Fig. 3-2 Diagram for swimming pool model's working mechanism |
A "swimming pool" chamber model would have a lager AHL concentration when compared with a "passage" chamber because the former one has a minor dilution. According to a published research that the oscillation period will be shorter as the AHL concentration increases. So we assume that it will be more time-consuming to use this "swimming pool" model in pretests.
However, this chamber is about 100 μm deep and could grow E. coli far more than monolayer. As time goes by, the oscillation in a single colony signaling by AHL could never go back to the original baseline, and the oscillation curves have a tendency like the following graph (Fig. 3-3).
 |
| Fig. 3-3 Simulation curve for our oscillation in "Swimming Pool" |
2.Advanced "Passage"
According to what we have discussed above, the microfluidic device should be able to provide flowing fresh media and maintain a constant bacteria population with single-cell resolution. Because "swimming pool" has a cell stacking problem, we designed a new passage-like device as shown below (Video 1).
Video 1
Channels and Chambers
This device is composed by two parts: the flowing channels and near 3,000 square trapping chambers, and their layout is like there are many passages with symmetry rooms distributed on both sides, so we call this array "passage" model. Firstly, cells are loaded from the cell port. When the loading stops, most cells would remain in channels, but if the loading cell density is dense enough, many cells would be pushed into chambers and form a monolayer of Escherichia coli growing in it. This is the time for running LB to get in. Fresh LB is loaded from the media port with Tween 20* and appropriate antibiotics. Flowing LB does not only permit a constant nutrition and exponentially growing cells for more than two days (that's the longest time we have tried), but also washes away newly generated cells to sustain a certain cell density, which is crucial for the determination of fluorescence strength.
Size
It has been stated before in the Mechanism part that most quorum sensing systems have a critical cell density requirement, so the trap size is designed neither too big nor too small to capture enough cells and allows for nutrients to permeate to every corner of the chamber.
You may also notice that the height of the chamber is extremely small, which is really demanding for the researcher's skills, but it's important for the observation. The oscillation could only be seen when the height of the chamber is in a particular range as our modeling predicted for many complicated reasons. For more details, please refer to the Modeling part.
Material
Polydimethylsiloxane (PDMS), a transparent polymer material, is chosen to build the devise because of its excellent breathability, which is necessary for the permeation of H2O2. Once H2O2 diffused outside cells, it can travel from colony to colony and synchronize them.
Our device is not only environmental friendly, but also a reusable one. After the flushing of a lysis buffer, most cells can be washed away, even those attached to walls. Then the device will be washed for three times by sterilized water before reuse.
 |
| Fig. 3-4 Microfluidic array- "Passage Model" |
Image Capturing & Data analysis
The green fluorescence of bacteria in chambers is captured every 5 minutes by an inverted microscope (Eclipse Ti, Nikon) with a standard CCD camera manually and usually last more than 7 hrs. (This is really a big challenge for our team and very physically demanding since this is the only available fluorescence we could find. However, we managed to finish this data collecting process anyway. Good jobs, guys!)
Data are transferred into oscillation curves by ImageJ and software designed by our teammate for digital image batch processing. (More detail for our self-designed software, please refer to protocol.)
 | Fig. 3-4 Microfluidic array- "Passage Model" |
Data are transferred into oscillation curves by ImageJ and software designed by our teammate for digital image batch processing. (More detail for our self-designed software, please refer to Software.
Next, we are going to Improvement (For a better Glee) to see how our little friends do on stage.
Mathematical model
We modeled the oscillate gene circuit (without ndh gene) based on the paper "A synchronized quorum of genetic clocks"[12]. In that, the author characterized the oscillator by using the four delay differential equations (DDEs), which are the basic of our modeling. DDE can perfectly simulate our circuit due to its powerful function about solving the dynamic change in biological processes.

In the first two equations, 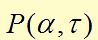 item is a Hill function equaling to
item is a Hill function equaling to  , which describes the delayed reactions about the complex processes in the cells (transcription, translation, maturation, etc.). This Hill function takes the history of the system into account, i.e. the concentration of AHL at the time it binds to LuxR to form the activation complex.
, which describes the delayed reactions about the complex processes in the cells (transcription, translation, maturation, etc.). This Hill function takes the history of the system into account, i.e. the concentration of AHL at the time it binds to LuxR to form the activation complex.
This model does not include an equation for LuxR assuming that it is constitutively produced at a constant level. All parameters are normalized for the convenience of analysis.
Modeling in MATLAB
We used MATLAB to help us solve the DDEs and draw the graphs. In addition, we developed a GUI user interface after finishing the main DDE code. The user interface makes people easier to modify and adjust the parameters without recoding the .m files and any MATLAB language knowledge. We called this software "Gene OS" which is short for Gene oscillate simulator.
See more in Software
After travelling a long way of circuit construction, you may wonder what we are going to do with those sparkling bacteria. Of course much thought was given, too, to make our oscillation circuit both showy and substantial as a safety guard in daily life. Here we are going to show you the potential of those shinny soldiers as biosensor, cancer cell killer and gene sinusoidal signal generator.
Biosensor
To date there appears a great number of biosensors that are used for detecting certain factors, such as small organic molecules or metal ions. And we've noticed that each year many iGEM teams relate their projects to biosensors. Biological detection has strength in its low detection limit, high sensitivity and bioaffinity when compared with chemical detection methods. We can use the same gene sensing circuit to detect various metal ions or other determinant via changing different promoters which responds to a specific target molecule.
Compared with traditional single-cell biosensor, our oscillatory system biosensor has three advantages.
1) Synchronized colonies will be more sensitive to concentration changes of the target molecule and the changes in fluorescence strength are easier to observe than single cell biosensors.
2) Single cell biosensor usually works, or "switches on" when the concentration of the specific molecules which can be responded by the promoter reaches to the lowest value. Sometimes this kind of detection is limited when we want to narrow the range of detection. Because the oscillation will happen only when concentration of certain molecule is between two value, the maximum and the minimum, it can dwindle the detection range of the determinant.
3) Oscillatory system is more robust and stable when used as a biosensor. These advantages will promote oscillatory gene circuits a better use in biosensing.
Jeff Hasty from UCSD used the same kind of oscillate circuit to engineer two kinds of arsenic-sensing macroscopic biosensors. These circuits are rewired by a native arsenite-responsive promoter, which is repressed by ArsR in the absence of arsenite. Due to the different mechanism, the sensor outputs are not the same. Figure 1 shows two constructed sensing modules and Figure 2 compares the outputs of two sensors[1].

Figure 1.Two different constructed sensing modules.


Figure 2. Two different outputs of corresponding biosensors.
Due to the accuracy and specific condition in the field of bio-medicine, the oscillatory biosensor needs to meet the requirement such as synchronization. For example, all cells expected to twinkle at the same time only when cytotoxin existed. In addition, the period or amplitude of the oscillation needs to indicate the concentration of the cytotoxin. Our circuit has the potential to accomplish this kind of job. By further exploration in the relationship between oscillatory parameters such as period and amplitude with other environmental factors such as concentration of antibiotic or cytotoxin, we may create a oscillatory biosensor which can sense different kinds of biological toxin.
Cancer cell killer
1.Cancer cell stalking
Nowadays cancer becomes one of the greatest enemy of our human being. Our main methods used to diagnosis of cancer are based on tissue and organ level, such as MRI, CT, PET, etc. These are mainly physical diagnostic methods.
With the development of science and technology, we create various methods into molecule level concentrated on molecular recognition and signal transduction of tumor marker, such as AFP(alpha fetoprotein), CEA (carcino-embryonic antigen), PSA(Prostate Specific Antigen) and so on. Level of these special molecules in human bodies has become the main basis of diagnosis of cancer. These are mainly chemical diagnostic methods.
So how about using synthetic biology to do something for diagnosis of cancer? Our project can be a potential biological method to assist diagnosis of cancer.
ROS is reactive oxygen species in cells, which has been proved to be associated with several neurological disorders including Alzheimer's disease (AD) and Parkinson's diseases[2][3][4]. Concentration of ROS indicates the level of cell metabolism. Normally the stronger cell' metabolism is, the more ROS it will produce. In the recent decade scientist found that the occurrence of cancer is closely related to the level of ROS. But it is not a simple positive correlation as we expect[5].

Figure 4. ROS Level
From this picture we can get to know it clearly. As we know, we use oxide stress to describe the level of ROS in our cell. There is a threshold of oxide stress which is the critical value between normal cell and cancer cell. As a normal cell, its oxide stress is lower than the critical value while the ROS production value is over the threshold. On the one hand, when the oxide stress in our internal environment increases, normal cells may mutate to cancer cells if the ROS value is higher than the threshold value. On the other hand, cancer cells may die because of the accumulation of ROS. So the key point is, when ROS value higher than the maximum, we can kill the cancer cell while ROS value lower than the threshold we may push normal cells to change into cancer cells. So the level of ROS is in a specific range which we can measure using chemical methods.
Think about a proper range of ROS's concentration. Is it familiar to you as we mentioned a similar problem when talking about advantages of oscillatory system as biosensors? H2O2 is a kind of ROS. The oscillation will happen only when the concentration of is H2O2 between the maximum and the minimum. By regulating our circuits to let it respond in the proper concentration range which matches the ROS range of cancer cells, we can use the oscillatory system as a biological cancer detector.

In our experiments, we found that we can regulate flow rate and some parameters of microfluidic chip such as the depth of the well, the width of the channel and the concentration of H2O2 to change the period and amplitude of the oscillation. So by changing parameters of microfluidic chip, we can get a proper concentration range of H2O2 if the oscillation works. From then on, we can extract some cancer tissue to detect the ROS concentration in the microfluidic chips or get some suspected tissue on chips and diagnose whether it contains cancer cell.
2 Periodical Killer
2.1 Drug delivery
Our circuit also has a great potential application in periodic drug delivery. It is amazing when we realize that the oscillate circuit can control the drug to release in a specific period and the concentration of the drug is related to the amplitude. It is well-known that nowadays drug-release is a very hot research field. Many efforts are focusing on how to modify or delivery drug using materials such as synthetic polymers, liposome or nano shell which can respond to differences between different intracellular environments. Our oscillatory gene circuits provide a more biofriendly way for drug release. Our future plan is to make some deep exploration in this field.
2.2 Protein Synthesis
What else, a resonance may happen among the oscillators which results in a peak synthesis of protein under some conditions. This phenomenon can be used to increase the yield of some medical proteins such as antitoxic serum and antibody[9][10].
2.3 Prevention pathogen
In the applications of controlling pathogens, quorum sensing system plays an important role in regulating growth and reputation of microorganism. So we could restrain quorum sensing system to control microorganism to make our life more healthy and nice. The most important part in the metabolism of pathogens controlled by quorum sensing system is the synthesis of toxin and the formation of membrane system. If we can restrain these two kinds of effect may we decrease some potential diseases resources and improve the effects of antitoxins. What's more, it is a particularly attractive point to prevent pathogens through restraining their quorum sensing system instead of killing them directly as it offers the prospect of combating effectively infections resistant to all existing drugs. As we all know, just treat diseases via using antibotics or antitoxin can help these pathogens improve their resistance to various drugs, which is very dangerous for us human being. To date, although we've found that different fungus form different signal molecules, the quorum sensing system almost takes part in cell morphological transformation and information communication. We think our oscillatory gene circuits provide a very useful and meaningful model to promote more and more research in this field[11].
Gene sinusoidal signal generator
With the development of science and technology, computer had been here and there with larger RAM, faster CPU and smaller volume. All these changes owe to VLSI (Very Large Scale Integrated Circuites), which consists of different electronic component.
Just image what if we can control cell as a biocomputer. Cellular computer is a device consists of different kinds genetically engineered bacteria, which can do the calculation, such as add and subtract like a normal computer. To finish such a device, scientists need various gene circuits to complete different operations. These circuits are just like electronic component working in an electronic computer.
At the beginning of this century, biocomputer is still a crazy dream far more than a possible blueprint. But now scientists have had the technology to make this "impossible mission" comes true. In recent months, scientists from Stanford constructed a new "bio-crystal valve" by DNA and RNA, which can do the logical operations in the living cells[6].
Two months later, scientists from MIT used less than 3 gene devices to do the digital operation in division, logarithmic and square root. Thus, constructing gene circuits as electronic devices have become possible. From their research, their cellular calculator which uses analog computation has more advantages in computing efficiency in part count, speed and energy consumption compared with digital computation[7].


Figure 1: Different kinds of log-domain analog computation.
Due to the fact that our circuits can generate a steady oscillation, it is possible that using our circuits as a sinusoidal signal generator.
Signal generators, such as function generator (including sinusoidal signal generator), are electronic devices that generate repeating or non-repeating electronic signals. As an important part of basic circuit studies, they are generally used in designing, testing, troubleshooting, and repairing electronic or electroacoustic devices.
What's more, Li Guang-bi from TUST reported that they have constructed two kinds of electronic circuits which well simulated the gene oscillator. This indicates that our circuit can develop to a new kind of gene sinusoidal signal generator[8].

Figure 2.This picture shows a kind of electronic circuit generating the oscillation wave.

Figure 3.This graph shows the generated oscillation wave.
Future Version Trilogy
We blueprint our next step developing oscillatory gene circuits in following steps:
Step One
Complete our project by cleaning every obstacle on our way to oscillation.
Confirm that every part is functioning as expected;
Amplify the effect of different copy numbers;
Complete the comparison of different strains' effect on oscillation;
Complete the comparison of different EGFP & sfGFP effect on oscillation;
Confirm most suitable microfluidic parameters for oscillation;
Optimize our model
Step Two
We will make our oscillation a controllable one by integrating the catalase gene into the circuit.
Catalase is a common enzyme found in nearly all living organisms exposed to oxygen. It catalyzes the decomposition of hydrogen peroxide to water and oxygen. It is a very important enzyme in protecting the cell from oxidative damage by reactive oxygen species (ROS). Since our lab is studying Shewanella oneidensis, and its catalase protein has showed outstanding performance in catalyzing H2O2. So we thought we could integrate the catalase gene into our circuit and under an artificial inducible promoter, by which we can decide when to start this catalase gene and stop the oscillation.
Step Three
Realize everything we imagined in APPLICATION!
Make our circuit function as biosensor, cancer cell killer and gene sinusoidal signal generator.
Reference
1. Prindle, A., et al., A sensing array of radically coupled genetic 'biopixels'. Nature 481, 39-44 (2012).
2. Gaggelli, E. Kozlowski, H. Valensin, D. Valensin & G. Copper ,Homeostasis and Neurodegenerative Disorders (Alzheimer's,Prion, and Parkinson's Diseases and Amyotrophic Lateral Sclerosis). Chem. Rev. 106, 1995–2044 (2006).
3. Scott, L. E. Orvig, C. Medicinal Inorganic Chemistry Approaches to Passivation and Removal of Aberrant Metal Ions in Disease. Chem. Rev. 109, 4885–4910 (2009).
4. Zheng, Z. Q. White, C. Lee, J. Peterson, T. S. Bush, A. I. Sun, G. Y. Weisman, G. A. & Petris, M. J. Altered Microglial Copper Homeostasis in a Mouse Model of Alzheimer's Disease. J. Neurochem. 114, 1630–1638 (2010).
5. Dunyaporn T., Jerome A. & Peng H., Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature Reviews Drug Discovery 8, 579-591 (2009).
6. Bonnet J, Yin P, Ortiz ME, Subsoontorn P & Endy D, Amplifying Genetic Logic Gates. Science 340, 599-603 ,2013.
7. Ramiz D., Timothy K. Lu et al. Synthetic analog computation in living cells. Nature 497, 619-+ (2013).
8. Guang bi Li et al, Electronic Ciircuit Model Construction of A Synthetic Gene Network. Journal of Tianjin University of Science & Technology 26, 22-25 (2011).
9. Hasty, J., Dolnik, M., Rottschafer, V. & Collins, J. J. Synthetic gene network for entraining and amplifying cellular oscillations. Physical Review Letters 88, 148101, (2002).
10. Elowitz, M. B. & Leibler, S. A. Synthetic oscillatory network of transcriptional regulators. Nature 403, 335 (2000).
11. Yan C., Quorum--Sensing System in Microbes and Its Application, School of Resources and Environment, Yuxi Normal University,Yuxi,Yunnan 653100.
12. Danino, T., Mondragon-Palomino, O., Tsimring, L. & Hasty, J., A synchronized quorum of genetic clocks. Nature 463, 326-330 (2010).
Parts
Table 1. The Bio-bricks used in the study
|
Part |
Backbone |
Type |
Location |
Size
(bp) |
Description |
|
|
part |
backbone |
|||||
|
BBa_K546000 |
pSB1C3 |
Signaling |
2013-P1-12D |
1964 |
2070 |
Lux pL
controlled LuxR with lux pR autoinducing LuxI (lva tag)- AHL. |
|
BBa_I763020 |
pSB1C3 |
Intermediate RBS-GFP-TT |
2013-P3-11H |
914 |
2070 |
RBS-GFP
(+LVA Tag)-Terminators |
|
BBa_F2621 |
pSB1A2 |
Signaling |
2013-P2-21F |
1158 |
2079 |
3OC6HSL
Receiver Device |
|
BBa_K546001 |
pSB1C3 |
Device |
2013-P1-12F |
2135 |
2070 |
AIIA AHL Reporter
and Quencher |
|
BBa_J04450 |
pSB4K5 |
Reporter |
2013-P5-5G |
1069 |
3419 |
RFP Coding
Device |
|
BBa_J04450 |
pSB3T5 |
Reporter |
2013-P5-7C |
1069 |
3241 |
RFP Coding Device |
|
BBa_J23022 |
BBa_J23006 |
Composite |
2013-P5-3H |
234 |
2356 |
[key3d][TT] |
| W/N | Name | Type | Description | Designer | Length |
|---|---|---|---|---|---|
| W | BBa_K1036000 | Composite | lux pL controlled luxR with lux pR controlled ndh (LVA-tag) coding for NADH dehydrogenase II | Shengquan Zeng Zhaopeng Cheng | 2687 |
| W | BBa_K1036001 | Coding | the ndh gene coding for respiratory NADH dehydrogenase II | Shengquan Zeng Zhaopeng Cheng | 1366 |
| W | BBa_K1036003 | Composite | lux pL controlled luxR with lux pR controlled gfp (LVA-tag) | Xin GuoXi Xi | 4052 |
| W | BBa_K1036006 | Composite | lux pL controlled luxR with lux pR controlled sfgfp (with LVA-tag) | Zhaopeng ChengXiyu Wu | 4046 |
| W/N | Name | Type | Description | Designer | Length |
|---|---|---|---|---|---|
| W | BBa_K1036002 | Composite | gfp (with LVA-tag) under plux R control | Xin Guo Xi Xi | 2866 |
| W | BBa_K1036004 | Reporter | sfgfp with LVA-tag | Zhaopeng ChengXiyu Wu | 753 |
| W | BBa_K1036005 | Composite | sfgfp (with LVA-tag) under plux R control | Zhaopeng ChengXiyu Wu | 2880 |




 "
"