Team:Heidelberg/Templates/DelH week17
From 2013.igem.org
(→Vector Maps and Primers) |
m |
||
| Line 56: | Line 56: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130822 BB16-17 G0 1kb+.png|200px|thumb|right|'''Fig.17.1''' gel of amplified DelH-G0 fragment from the fragment itself (loaded 20 µL of PCR) <br> ''l1:'' BB amplified with HM16 by 2-step PCR, ''l2:'' BB amplified with HM17by 2-step PCR, ''l3:'' BB amplified with HM16, ''l4:'' BB amplified with HM17, ''l5:'' DelH-G0 <br> l5: G0 was not amplified => not our fragment or without DMSO it is not so effective ]] | [[File:Heidelberg_20130822 BB16-17 G0 1kb+.png|200px|thumb|right|'''Fig.17.1''' gel of amplified DelH-G0 fragment from the fragment itself (loaded 20 µL of PCR) <br> ''l1:'' BB amplified with HM16 by 2-step PCR, ''l2:'' BB amplified with HM17by 2-step PCR, ''l3:'' BB amplified with HM16, ''l4:'' BB amplified with HM17, ''l5:'' DelH-G0 <br> l5: G0 was not amplified => not our fragment or without DMSO it is not so effective ]] | ||
| - | + | ||
Gel does not show expected band. | Gel does not show expected band. | ||
:=> G0 was not amplified. Possibly change Mg<sup>2+</sup> concentration. | :=> G0 was not amplified. Possibly change Mg<sup>2+</sup> concentration. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
====PCR Conditions G0.W17.B==== | ====PCR Conditions G0.W17.B==== | ||
{| class="wikitable" style="float:left; margin-right:1em" | {| class="wikitable" style="float:left; margin-right:1em" | ||
| Line 107: | Line 108: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130823 G00,5 G0-1 G0-1,5 GoN.png|200px|thumb|right|'''Fig.17.2''' gel of amplified DelH-G0 with different MgSO<sub>4</sub>(loaded 20 µL of PCR) <br> ''l1:'' 1Kb+ ladder, ''l2:'' G0 0.5 µl MgSO<sub>4</sub>, ''l3:'' G0 1 µl MgSO<sub>4</sub>, ''l4:'' G0 1.5µl MgSO<sub>4</sub>, ''l5:'' G0 N without MgSO<sub>4</sub> <br> l5: G0 was amplified without DMSO => DMSO is important, but MgSO<sub>4</sub> is not improve the PCR reaction]] | [[File:Heidelberg_20130823 G00,5 G0-1 G0-1,5 GoN.png|200px|thumb|right|'''Fig.17.2''' gel of amplified DelH-G0 with different MgSO<sub>4</sub>(loaded 20 µL of PCR) <br> ''l1:'' 1Kb+ ladder, ''l2:'' G0 0.5 µl MgSO<sub>4</sub>, ''l3:'' G0 1 µl MgSO<sub>4</sub>, ''l4:'' G0 1.5µl MgSO<sub>4</sub>, ''l5:'' G0 N without MgSO<sub>4</sub> <br> l5: G0 was amplified without DMSO => DMSO is important, but MgSO<sub>4</sub> is not improve the PCR reaction]] | ||
| - | + | ||
Fragments were amplified, but yield not significantly increased. | Fragments were amplified, but yield not significantly increased. | ||
:=> DMSO is important, but MgSO<sub>4</sub> is not improve the PCR reaction. | :=> DMSO is important, but MgSO<sub>4</sub> is not improve the PCR reaction. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
===Characterization of DelH Plasmid pHM03 17-08=== | ===Characterization of DelH Plasmid pHM03 17-08=== | ||
* For the miniprep, the cells were centrifuged at 13,000 rpm for 15 min | * For the miniprep, the cells were centrifuged at 13,000 rpm for 15 min | ||
| Line 186: | Line 188: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130818_Colony_PCR_DelH.png|200px|thumb|right|'''Fig.17.3''' colony PCR of Gibson assembled DelH-BB with mRFP (loaded 20 µL of PCR) <br> ''l1:'' <br> ''l3-12'':show expected fragment ]] | [[File:Heidelberg_20130818_Colony_PCR_DelH.png|200px|thumb|right|'''Fig.17.3''' colony PCR of Gibson assembled DelH-BB with mRFP (loaded 20 µL of PCR) <br> ''l1:'' <br> ''l3-12'':show expected fragment ]] | ||
| - | + | ||
Colonies 3 to 12 show expected size. | Colonies 3 to 12 show expected size. | ||
:=> Perform miniprep and test digest with colonies 3-12. | :=> Perform miniprep and test digest with colonies 3-12. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
====Test Restriction Digest==== | ====Test Restriction Digest==== | ||
Of colonies 3, 4, 5, 6, 7 | Of colonies 3, 4, 5, 6, 7 | ||
| Line 211: | Line 214: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130819 digestedColonies3-7.png|200px|thumb|right|'''Fig.17.4''' gel of test digest of the colonies 3-7 (loaded 20 µL of PCR) <br> ''l1:''1 Kb+ ladder, ''l2:''Digested miniprep of colony 3, ''l3:''Digested miniprep of colony 4, ''l4:''Digested miniprep of colony 5, ''l5:''Digested miniprep of colony 6, ''l6:''Digested miniprep of colony 7 <br> ''l2-6'':show not the expected fragment, but a band at ~ 4.8 Kb. Also observed pale bands at 20 Kb and ~ 3.8 Kb]] | [[File:Heidelberg_20130819 digestedColonies3-7.png|200px|thumb|right|'''Fig.17.4''' gel of test digest of the colonies 3-7 (loaded 20 µL of PCR) <br> ''l1:''1 Kb+ ladder, ''l2:''Digested miniprep of colony 3, ''l3:''Digested miniprep of colony 4, ''l4:''Digested miniprep of colony 5, ''l5:''Digested miniprep of colony 6, ''l6:''Digested miniprep of colony 7 <br> ''l2-6'':show not the expected fragment, but a band at ~ 4.8 Kb. Also observed pale bands at 20 Kb and ~ 3.8 Kb]] | ||
| - | + | ||
None of the colonies shows expected restriction fragments. Additionally, there are pale bands at +20 Kb and ~3.8 Kb, but not the expected bands. | None of the colonies shows expected restriction fragments. Additionally, there are pale bands at +20 Kb and ~3.8 Kb, but not the expected bands. | ||
:=> Possible explanation: colonies harbor reconjugated backbone AND something else involving DelH. | :=> Possible explanation: colonies harbor reconjugated backbone AND something else involving DelH. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
====PCR Conditions CP.W17.B==== | ====PCR Conditions CP.W17.B==== | ||
M3 = Miniprep of DelH-pSB6A1(colony 3) | M3 = Miniprep of DelH-pSB6A1(colony 3) | ||
| Line 258: | Line 262: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130820_col_PCR_delH_zoom.png|200px|thumb|right|'''Fig.17.5''' colony PCR of Gibson assembled DelH-BB with mRFP (loaded 20 µL of PCR) <br> ''l1:'' <br> ''l3-12'':show an unexpected fragment at ~ 3Kb ]] | [[File:Heidelberg_20130820_col_PCR_delH_zoom.png|200px|thumb|right|'''Fig.17.5''' colony PCR of Gibson assembled DelH-BB with mRFP (loaded 20 µL of PCR) <br> ''l1:'' <br> ''l3-12'':show an unexpected fragment at ~ 3Kb ]] | ||
| - | + | ||
Gel shows unexpected fragments at ~3 Kb. | Gel shows unexpected fragments at ~3 Kb. | ||
:=> Clones are negative despite of latest results. Current strategy does not work out. '''Development new strategy.''' | :=> Clones are negative despite of latest results. Current strategy does not work out. '''Development new strategy.''' | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
===New Gibson Strategy=== | ===New Gibson Strategy=== | ||
====Gibson Strategy without mRFP==== | ====Gibson Strategy without mRFP==== | ||
| Line 346: | Line 351: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130822 tetR1-2.png|200px|thumb|right|'''Fig.17.6''' gel of amplified tetracycline resistance (loaded 20 µL of PCR) <br> ''l1:'' 2 log, ''l2:'' tetracycline resistance amplified from colony, ''l3:'' tetracycline resistance amplified from colony 2 <br> l2-3: show no band = maybe annealing temperature to high]] | [[File:Heidelberg_20130822 tetR1-2.png|200px|thumb|right|'''Fig.17.6''' gel of amplified tetracycline resistance (loaded 20 µL of PCR) <br> ''l1:'' 2 log, ''l2:'' tetracycline resistance amplified from colony, ''l3:'' tetracycline resistance amplified from colony 2 <br> l2-3: show no band = maybe annealing temperature to high]] | ||
| - | + | ||
Gel does not schow any band. | Gel does not schow any band. | ||
:=> Try amplification with lower annealing temperature. | :=> Try amplification with lower annealing temperature. | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
====PCR Conditions TR.W17.B==== | ====PCR Conditions TR.W17.B==== | ||
{| class="wikitable" style="float:left; margin-right:1em" | {| class="wikitable" style="float:left; margin-right:1em" | ||
| Line 395: | Line 400: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130823 tetR 2log.png|200px|thumb|right|'''Fig.17.7''' gel of amplified tetracycline resistance (loaded 20 µL of PCR) <br> ''l1:'' 2 log, ''l2:'' tetracycline resistance amplified from new picked colony 1, ''l3:'' tetracycline resistance amplified from new picked colony 2, <br> l2-3: show no band]] | [[File:Heidelberg_20130823 tetR 2log.png|200px|thumb|right|'''Fig.17.7''' gel of amplified tetracycline resistance (loaded 20 µL of PCR) <br> ''l1:'' 2 log, ''l2:'' tetracycline resistance amplified from new picked colony 1, ''l3:'' tetracycline resistance amplified from new picked colony 2, <br> l2-3: show no band]] | ||
| - | + | ||
Gel does not show expected band. | Gel does not show expected band. | ||
:=> Repeat amplification with miniprep, therefore miniprep the 3 colonies containing pSB1T3. | :=> Repeat amplification with miniprep, therefore miniprep the 3 colonies containing pSB1T3. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
====PCR Conditions TR.W17.C==== | ====PCR Conditions TR.W17.C==== | ||
{| class="wikitable" style="float:left; margin-right:1em" | {| class="wikitable" style="float:left; margin-right:1em" | ||
| Line 442: | Line 448: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130823 tetR 2log.png|200px|thumb|right|'''Fig.17.8''' gel of amplified tetracycline resistance (loaded 20 µL of PCR) <br> ''l1:'' tetracycline resistance amplified from minipreped colony 1, ''l2:'' tetracycline resistance amplified from minipreped colony 2, ''l3:'' tetracycline resistance amplified from minipreped colony 3,''l4:'' 2 log, <br> l1-3: show expected band = was cut]] | [[File:Heidelberg_20130823 tetR 2log.png|200px|thumb|right|'''Fig.17.8''' gel of amplified tetracycline resistance (loaded 20 µL of PCR) <br> ''l1:'' tetracycline resistance amplified from minipreped colony 1, ''l2:'' tetracycline resistance amplified from minipreped colony 2, ''l3:'' tetracycline resistance amplified from minipreped colony 3,''l4:'' 2 log, <br> l1-3: show expected band = was cut]] | ||
| - | + | ||
Gel does show expected band. | Gel does show expected band. | ||
:=> Fragment was cut and gel extracted (c= 132.1 ng/µl). | :=> Fragment was cut and gel extracted (c= 132.1 ng/µl). | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
===Amplification of Backbone=== | ===Amplification of Backbone=== | ||
====PCR Conditions BB.W7.A==== | ====PCR Conditions BB.W7.A==== | ||
| Line 491: | Line 497: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130821 BB-PCCR.png|200px|thumb|right|'''Fig.17.9''' gel of amplified BB (loaded 25 µL of PCR) <br> ''l1:'' BB amplified with HM16, ''l2:'' BB amplified with HM17 <br> l1:specific band as expected = was cut out]] | [[File:Heidelberg_20130821 BB-PCCR.png|200px|thumb|right|'''Fig.17.9''' gel of amplified BB (loaded 25 µL of PCR) <br> ''l1:'' BB amplified with HM16, ''l2:'' BB amplified with HM17 <br> l1:specific band as expected = was cut out]] | ||
| - | + | ||
Gel shows specific band for amplification using HM16. | Gel shows specific band for amplification using HM16. | ||
:=> Fragment wa scut and gel extracted. For HM17, run a two-step PCR because of it's high annealing temperature. Also we are going to rise the amplification time to 1:30 min. | :=> Fragment wa scut and gel extracted. For HM17, run a two-step PCR because of it's high annealing temperature. Also we are going to rise the amplification time to 1:30 min. | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
====PCR Conditions BB.W7.B==== | ====PCR Conditions BB.W7.B==== | ||
{| class="wikitable" style="float:left; margin-right:1em" | {| class="wikitable" style="float:left; margin-right:1em" | ||
| Line 538: | Line 544: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130822 BB16-17 G0 1kb+.png|200px|thumb|right|'''Fig.17.10''' gel of amplified BB with different primers (loaded 20 µL of PCR) <br> ''l1:'' BB amplified with HM16 by 2-step PCR, ''l2:'' BB amplified with HM17by 2-step PCR, ''l3:'' BB amplified with HM16, ''l4:'' BB amplified with HM17, ''l5:'' DelH-G0 <br> all 4 samples (l1-4) showed a specific band at 4.4 Kb = was cut]] | [[File:Heidelberg_20130822 BB16-17 G0 1kb+.png|200px|thumb|right|'''Fig.17.10''' gel of amplified BB with different primers (loaded 20 µL of PCR) <br> ''l1:'' BB amplified with HM16 by 2-step PCR, ''l2:'' BB amplified with HM17by 2-step PCR, ''l3:'' BB amplified with HM16, ''l4:'' BB amplified with HM17, ''l5:'' DelH-G0 <br> all 4 samples (l1-4) showed a specific band at 4.4 Kb = was cut]] | ||
| - | + | ||
All samples show expected length. | All samples show expected length. | ||
:=> Fragments were cut and gel extracted. | :=> Fragments were cut and gel extracted. | ||
| Line 550: | Line 556: | ||
|} | |} | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
===Restriction Digest with DpnI=== | ===Restriction Digest with DpnI=== | ||
* Incubated at 37°C for 2.5 h | * Incubated at 37°C for 2.5 h | ||
| Line 656: | Line 662: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130823 2log screening1-4end.svg.png|200px|thumb|right|'''Fig.17.11''' colony PCR of Gibson assembled DelH-BB with mRFP (loaded 20 µL of PCR) <br> ''l1:'' <br> ''l3-12'':show an unexpected fragment at ~ 3Kb ]] | [[File:Heidelberg_20130823 2log screening1-4end.svg.png|200px|thumb|right|'''Fig.17.11''' colony PCR of Gibson assembled DelH-BB with mRFP (loaded 20 µL of PCR) <br> ''l1:'' <br> ''l3-12'':show an unexpected fragment at ~ 3Kb ]] | ||
| - | + | ||
Gel does not show expected bands. | Gel does not show expected bands. | ||
:=> Screen 10 minipreped colonies of the plate 5 and pick another 10 colonies of plate 1, 2, 3 and 6 (on plate 4 are no colonies). | :=> Screen 10 minipreped colonies of the plate 5 and pick another 10 colonies of plate 1, 2, 3 and 6 (on plate 4 are no colonies). | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
====Colony-PCR CP.W17.D==== | ====Colony-PCR CP.W17.D==== | ||
* 2 different screening are performed | * 2 different screening are performed | ||
| Line 707: | Line 713: | ||
Expected band: 663 bp (screening start), 2.5 Kb (screening end) | Expected band: 663 bp (screening start), 2.5 Kb (screening end) | ||
<br/> | <br/> | ||
| - | [[File: | + | [[File:Heidelberg_20130826 2log screening 1F-6J.png|200px|thumb|right|'''Fig.17.13''' colony PCR of Gibson assembled DelH-BB without mRFP with the screening primer VR2 and HM13 (loaded 1 µL of PCR) <br> ''l1:''2log ladder ''l2-24:'' PCR of picked colonies from plate 1,2,3,5,6 <br> no band at 663 bp observed]] |
| - | with the screening primer | + | <div class="tright" style="clear:none">[[File:Heidelberg_20130824 2log screening 1A-6E.png|200px|thumb|right|'''Fig.17.12''' colony PCR of Gibson assembled DelH-BB without mRFP |
| - | + | with the screening primer VF2 and DN11(loaded 1 µL of PCR) <br> ''l1:''2log ladder ''l2-24:'' PCR of picked colonies from plate 1,2,3,5,6 <br> no band at 663 bp observed]]</div> | |
| - | + | ||
None of the gels shows expected fragment. | None of the gels shows expected fragment. | ||
:=> Overthink strategy. | :=> Overthink strategy. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
Latest revision as of 13:48, 25 October 2013
Contents
|
19-08 - 25-08-13
Summary of Amplified DelH Fragments
| Fragment | Concentration [ng/µl] |
|---|---|
| G0 | 6.4 |
| G1/2a | 8.5 |
Rather low yield. So we switched the strategy and try to increase the template of DelH by amplifying it in smaller and more fragments. Therefore, we used the following primers: FS64-FS71, HM08, DN07
Amplification of DelH G0
Re-PCR Conditions G0.W17.A
| Reagent | G0 |
|---|---|
| Expected length [Kb] | 18 |
| Named | G0 1 |
| Template | 1 µl G0 fragment (c= 6.4 ng/µl) |
| Primer fw 10 µM | 2 µl short2 |
| Primer rev 10 µM | 2 µl HM08 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb

l1: BB amplified with HM16 by 2-step PCR, l2: BB amplified with HM17by 2-step PCR, l3: BB amplified with HM16, l4: BB amplified with HM17, l5: DelH-G0
l5: G0 was not amplified => not our fragment or without DMSO it is not so effective
Gel does not show expected band.
- => G0 was not amplified. Possibly change Mg2+ concentration.
PCR Conditions G0.W17.B
| Reagent | G0 | G0 | G0 | G0 |
|---|---|---|---|---|
| Expected length [Kb] | 18 | 18 | 18 | 18 |
| Named | G0 0.5 | G0 1 | G0 2 | G0 N |
| Template | 1 µl glycerol stock D. acidovorans | 1 µl glycerol stock D. acidovorans | 1 µl glycerol stock D. acidovorans | 1 µl glycerol stock D. acidovorans |
| Primer fw 10 µM | 2 µl short2 | 2 µl short2 | 2 µl short2 | 2 µl short2 |
| Primer rev 10 µM | 2 µl HM08 | 2 µl HM08 | 2 µl HM08 | 2 µl HM08 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 4.5 µl | 4 µl | 3.5 µl | 4 µl |
| DMSO | 1 | 1 | 1 | 1 |
| MgSO4 | 0.5 µl | 1 µl | 1.5 µl | - |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:45 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Lid preheated at 98°C
- No hot start
Result
Expected band: 18 Kb
Fragments were amplified, but yield not significantly increased.
- => DMSO is important, but MgSO4 is not improve the PCR reaction.
Characterization of DelH Plasmid pHM03 17-08
- For the miniprep, the cells were centrifuged at 13,000 rpm for 15 min
- The cell pellet was resuspended in 200 µl P1 + 400 µl P2 and incubated for 120
- Afterwards, 300 µl S3 were added and transferred into a new 2 ml eppi and centrifuged 20 min at 13,000 rpm
- 1 ml isopropanol was added and the normal isoprop-ethanol-preipitation
- It was resuspended in 35 µl ddH2O
Result
| Miniprep of colony | Concentration [ng/µl] |
|---|---|
| 3 | 1,868.5 |
| 4 | 1,688.1 |
| 5 | 1,069.2 |
| 6 | 1,778.6 |
| 7 | 2,105.7 |
| 8 | 1,219.7 |
| 9 | 1,083.1 |
| 10 | 1,031.7 |
| 11 | 2,175.3 |
| 12 | 1,449.4 |
PCR Conditions CP.W17.A
M3 = Miniprep of DelH-pSB6A1(colony 3)
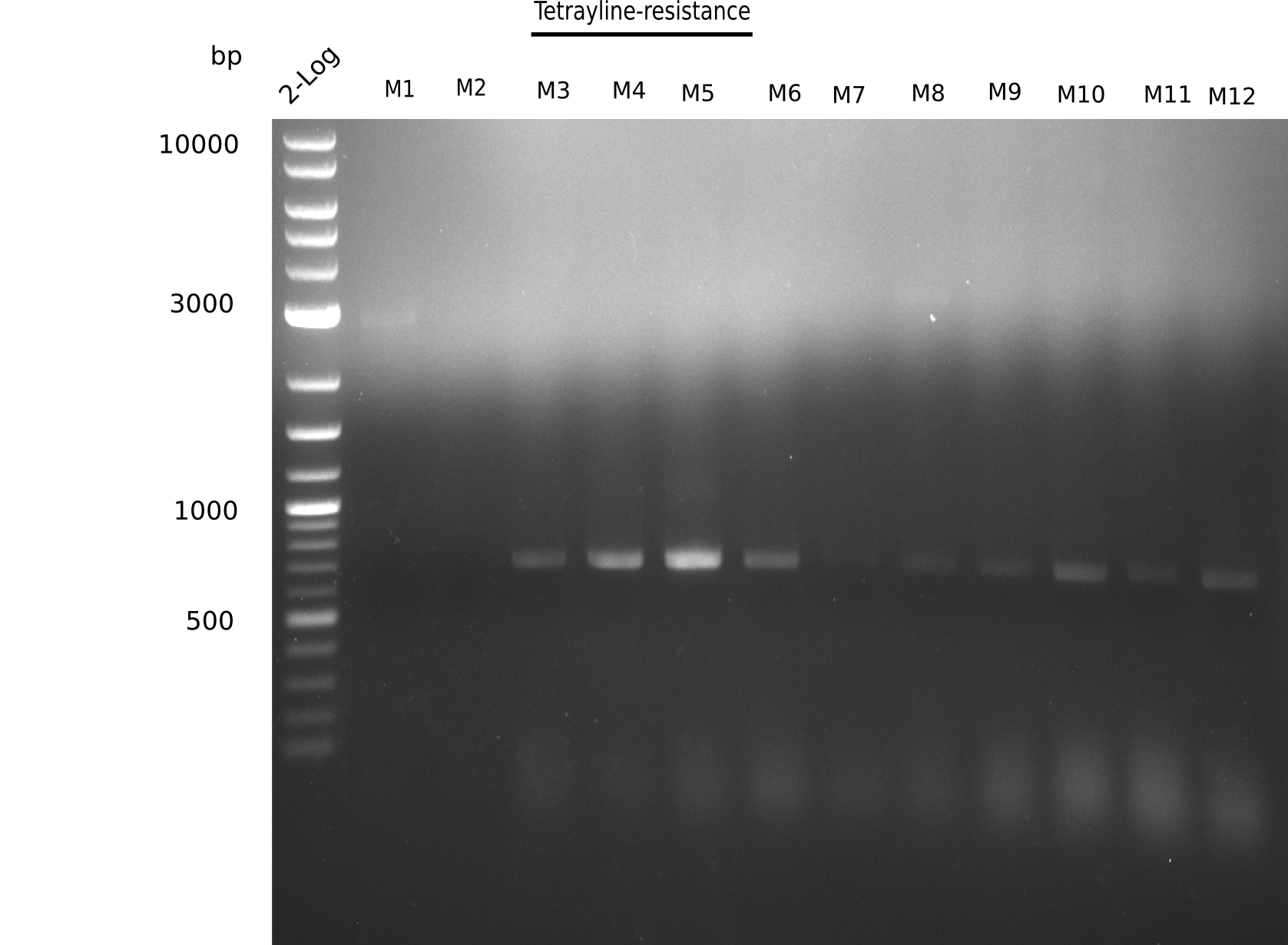
| Template | 1 µl of 1:1000 diluted M3 (pink) | 1 µl of 1:1000 diluted M4 (slightly pink) | 1 µl of 1:1000 diluted M5 (slightly pink) | 1 µl of 1:1000 diluted M6 (pink) | 1 µl of 1:1000 diluted M7 (pink) | 1 µl of 1:1000 diluted M 8 | 1 µl of 1:1000 diluted M 9 | 1 µl of 1:1000 diluted M10 | 1 µl of 1:1000 diluted M11 | 1 µl of 1:1000 diluted M12 |
|---|---|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature[°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Colonies 3 to 12 show expected size.
- => Perform miniprep and test digest with colonies 3-12.
Test Restriction Digest
Of colonies 3, 4, 5, 6, 7
| Component | Amount [µl] |
|---|---|
| DNA of M 3 to 12 (1:10) | 19 |
| CutSmart Buffer (10x) | .,2 |
| Enzyme (SalI) | 1 |
| ddH2O | - |
| Expected bands | 14,180 & 9,469 bp |
- Incubated over night at 37°C and shaking
Result
Expected bands: 14.18 and 9.459 Kb

l1:1 Kb+ ladder, l2:Digested miniprep of colony 3, l3:Digested miniprep of colony 4, l4:Digested miniprep of colony 5, l5:Digested miniprep of colony 6, l6:Digested miniprep of colony 7
l2-6:show not the expected fragment, but a band at ~ 4.8 Kb. Also observed pale bands at 20 Kb and ~ 3.8 Kb
None of the colonies shows expected restriction fragments. Additionally, there are pale bands at +20 Kb and ~3.8 Kb, but not the expected bands.
- => Possible explanation: colonies harbor reconjugated backbone AND something else involving DelH.
PCR Conditions CP.W17.B
M3 = Miniprep of DelH-pSB6A1(colony 3)
| Template | 1 µl of 1:1000 diluted M3 (pink) | 1 µl of 1:1000 diluted M4 (slightly pink) | 1 µl of 1:1000 diluted M5 (slightly pink) | 1 µl of 1:1000 diluted M6 (pink) | 1 µl of 1:1000 diluted M7 (pink) | 1 µl of 1:1000 diluted M 8 | 1 µl of 1:1000 diluted M 9 | 1 µl of 1:1000 diluted M10 | 1 µl of 1:1000 diluted M11 | 1 µl of 1:1000 diluted M12 |
|---|---|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Gel shows unexpected fragments at ~3 Kb.
- => Clones are negative despite of latest results. Current strategy does not work out. Development new strategy.
New Gibson Strategy
Gibson Strategy without mRFP
Try a new construct without mRFP, so that we can exclude the red clones from the screening. Therefore we are going to use a new reverse primer for the Backbone which includes only the terminator of the mRFP, but not the mRFP. The primers for the Backbone are HM11 & HM17. The construct is named pHM04 and shown in week 16 were the idea was specified.
For first approach pHM04, we will amplify the backbone without mRFP, avoiding backbone reassembly due to ribosome binding site homology. In the second strategy pHM05, we will additionally introduce a tetracycline resistance, to ensure integration of the insert.
Vector Maps and Primers
1st strategy: DelH & pSB6A1 without mRFP
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| HM17:DelH_Terminator_BB_fw | 16-08-2013 | Amplification of Backbone pSb6A1-lacI-mRFP | ATTGGCGCTGGAGTACGCGCTGGACTGA aggcatcaaataaaacgaaaggctcag |
2nd strategy: DelH & tetracycline resistance & pSB6A1 without mRFP
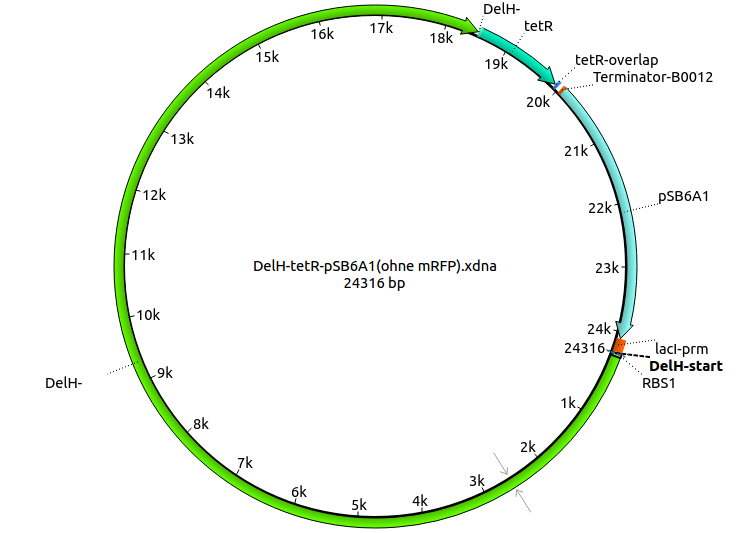
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| HM14:DelH_tetR_fw | 2013-08-16 | Gibson-Primer DelH-tetR: amplifies the tetracycline resistance from the pSB1T3 Backbone and creates an overlap to the end of DelH | ATTGGCGCTGGAGTACGCGCTGGACTGA atgaagttttaaatcaatctaaag |
| HM15:tetR_stop_BB_rev | 2013-08-16 | Gibson-Primer tetR-pSB6A1: amplifies the tetracycline resistance and creates an overlap with the Terminator of the Backbone pSB6A1 | Cgactgagcctttcgttttatttgatgcctggc ctcgtgatacgcctatttttatagg |
| HM16:tetR_pSB6A1_fw | 2013-08-16 | Gibson-Primer DelH, amplifies the Backbone pSB6A1 creating an overlap with the tetracycline resistance | Aaaaataggcgtatcacgag gccaggcatcaaataaaacgaaaggctcag |
Amplification of Tetracycline Fragment
Preparation of Tetraycline Medium
- Weight 100 mg of tetracycline and dissolve it in 10 ml ethanol = stock solution
- Add 500 µl of stock solution in 100 ml LB and mix
Transformation of Tetrazyklin Backbone
- For the new strategy we need a tetracycline resistance (length ~1.2 Kb) of the pSB1T3 Backbone (2013 Distribution, Plate 5, well 7A, insert: BBa_J04450) [http://parts.igem.org/Part:pSB1T3:Design|pSB1T3]
- The primers are already designed and shown in the table above
- With these primers, we amplify the tetracycline resistance and include an overlap to DelH end and the backbone pSB6A1 excluding the mRFP
- For the transformation I incubated 10 min at room temperature the well 7A of plate 5 with 10 µl ddH2O
PCR Conditions TR.W17.A
| Reagent | pSB1T3 | pSB1T3 |
|---|---|---|
| Template | picked colony 1 | picked colony 2 |
| Primer fw 10 µM | 1 µl HM14 | 1 µl HM14 |
| Primer rev 10 µM | 1 µl HM15 | 1 µl HM15 |
| Phusion Ready Mix | 10 µl | 10 µl |
| ddH2O | 8 µl | 8 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0,5°C) | 5 | |
| 72 | 30 | |
| 18 | 98 | 1 |
| 67 | 5 | |
| 72 | 30 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 1.468 Kb
Gel does not schow any band.
- => Try amplification with lower annealing temperature.
PCR Conditions TR.W17.B
| Reagent | pSB1T3 |
|---|---|
| Template | picked colony |
| Primer fw 10 µM | 1 µl HM14 |
| Primer rev 10 µM | 1 µl HM15 |
| Phusion Ready Mix | 10 µl |
| ddH2O | 8 µl |
| DMSO | - |
| Cycles | Temperature DelH BB [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 55 (touchdown -0,5°C) | 5 | |
| 72 | 30 | |
| 18 | 98 | 1 |
| 55 | 5 | |
| 72 | 30 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 1.468 Kb
Gel does not show expected band.
- => Repeat amplification with miniprep, therefore miniprep the 3 colonies containing pSB1T3.
PCR Conditions TR.W17.C
| Reagent | pSB1T3 | pSB1T3 | pSB1T3 |
|---|---|---|---|
| Template | Minipreped pSB1T3 | Minipreped pSB1T3 | Minipreped pSB1T3 |
| Primer fw 10 µM | 2 µl HM14 | 2 µl HM14 | 2 µl HM14 |
| Primer rev 10 µM | 2 µl HM15 | 2 µl HM15 | 2 µl HM15 |
| Phusion Ready Mix | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 55 (touchdown -0,5°C) | 5 | |
| 72 | 30 | |
| 18 | 98 | 1 |
| 55 | 5 | |
| 72 | 30 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: 1.468 Kb

l1: tetracycline resistance amplified from minipreped colony 1, l2: tetracycline resistance amplified from minipreped colony 2, l3: tetracycline resistance amplified from minipreped colony 3,l4: 2 log,
l1-3: show expected band = was cut
Gel does show expected band.
- => Fragment was cut and gel extracted (c= 132.1 ng/µl).
Amplification of Backbone
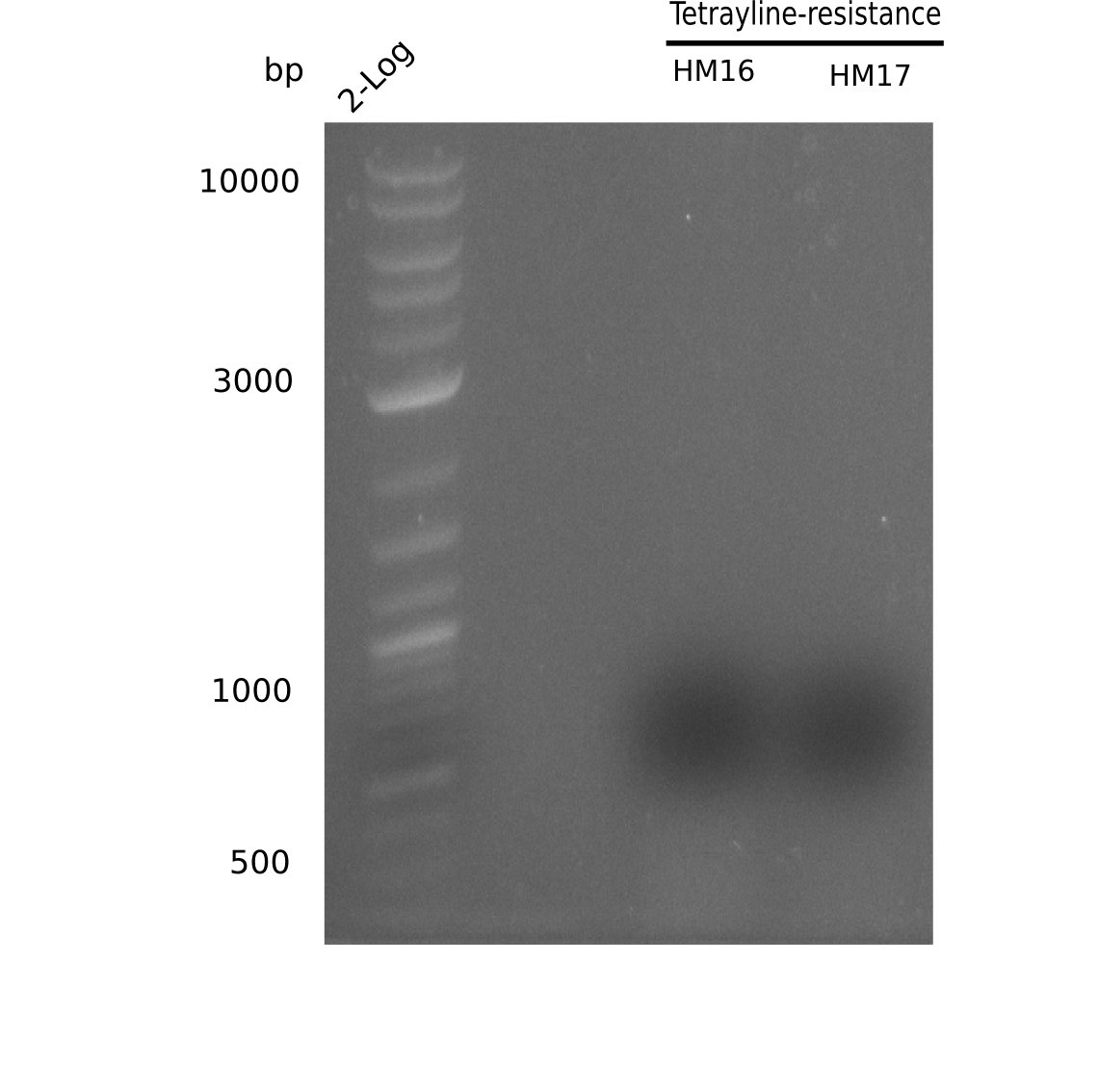
PCR Conditions BB.W7.A
With new primers (HM16 & HM17)
| Reagent | BB with tetracycline & without mRFP | BB without mRFP |
|---|---|---|
| Template | 1 µl BB 3.08 | 1 µl BB 3.08 |
| Primer fw 10 µM | 2 µl HM11 | 2 µl HM11 |
| Primer rev 10 µm | 2 µl HM16 | 2 µl HM17 |
| Phusion Flash Ready Mix | 10 µl | 10 µl |
| ddH2O 5µl | 5 µl |
| Cycles | Temperature DelH BB [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 69 (touchdown -0,5°C) | 5 | |
| 72 | 75 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 75 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: ~ 4.4 Kb
Gel shows specific band for amplification using HM16.
- => Fragment wa scut and gel extracted. For HM17, run a two-step PCR because of it's high annealing temperature. Also we are going to rise the amplification time to 1:30 min.
PCR Conditions BB.W7.B
| Reagent | BB with tetracycline & without mRFP | BB without mRFP | BB with tetracycline & without mRFP | BB without mRFP |
|---|---|---|---|---|
| Template | 1 µl BB 3,08 | 1 µl BB 3,08 | 1 µl BB 3,08 | 1 µl BB 3,08 |
| Primer fw 10 µM | 2 µl HM11 | 2 µl HM11 | 5 µl HM11 | 5 µl HM11 |
| Primer rev 10 µM | 2 µl HM16 | 2 µl HM17 | 5 µl HM16 | 5 µl HM17 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 25 µl | 25 µl |
| ddH2O | 5 µl | 5 µl | 14 µl | 14 µl |
| Cycles | Temperature (20 µl = sample 1 & 2) [°C] | Time | Temperature (50 µl = sample 3 & 4) [°C] | Time [s] |
|---|---|---|---|---|
| 1 | 98 | 5 | 98 | 5 |
| 12 | 98 | 1 | 98 | 1 |
| 69 (touchdown -0,5°C) | 5 | - | ||
| 72 | 90 | 72 | 90 | |
| 18 | 98 | 1 | 98 | 1 |
| 68 | 5 | - | ||
| 72 | 90 | 72 | 90 | |
| 1 | 72 | 5 min | 72 | 5 min |
| 1 | 4 | inf | 4 | inf |
Result
Expected band: ~ 4.4 Kb
All samples show expected length.
- => Fragments were cut and gel extracted.
| Sample | Concentration [ng/µl] |
|---|---|
| BB-16 | 76.7 |
| BB-17 | 51.3 |
Restriction Digest with DpnI
- Incubated at 37°C for 2.5 h
| Sample [µl] | Buffer [µl] | Enzyme [µl] |
|---|---|---|
| 18 BB-16 | 2.1 CutSmart Buffer | 1 DpnI |
| 18 BB-16 | 2.1 CutSmart Buffer | 1 DpnI |
- Afterwards, a purification was performed with the nucleotide removal kit
Generation of DelH Plasmid pHM04 23-08
Summary of Fragments
| Fragment | Concentration [ng/µl] | Date |
|---|---|---|
| G0 | 6.4 | 22-08 |
| G1/2a | 8.5 | 22-08 |
| G2b | 18.6 | Example |
| BB | Example | 22-08 |
Gibson Assembly
| Mix name | DelH G0 | DelH G1/2a | DelH G2b | BB17 | Gibson Master Mix [µl] | Final volume [µl] |
|---|---|---|---|---|---|---|
| E1 | 9 | - | - | 0.5 | 10 | 20 |
| E2 | - | 7 | 2.5 | 0.5 | 10 | 20 |
- Incubate the Gibson Assembly for 1 h at 50°C
Electroporation
| Sample | Electroporated with | Plated out | Cuvette "exploded" |
|---|---|---|---|
| 1 | 1 µl of E1 (5 µl Gibson + 10 µl ddH2O) | 22.08.2013 | v |
| 2 | 14 µl of E1 (5 µl Gibson + 10 µl ddH2O) | 22.08.2013 | v |
| 3 | 1 µl of E2 (5 µl Gibson + 10 µl ddH2O) | 22.08.2013 | - |
| 4 | 14 µl of E2 (5 µl Gibson + 10 µl ddH2O) | 22.08.2013 | v |
| 5 | 20 µl of E1 (purified by isoprop-precipitation) | 22.08.2013 | - |
| 6 | 20 µl of E2 (purified by isoprop-precipitation) | 22.08.2013 | - (no pellet observed) |
Colony-PCR CP.W17.C
- 50 colonies of plate 5 (= isoprop purified E1 => G0-complete in Backbone)
- 10 colonies of plate ...
- 2 different screening are performed
- PC = picked colony
- S5 = Sample 5 (watch names above on construct DelH-BB)
| Template | 50 x 1 PC S5 | 50 x 1 PC S5 | 4 x 1 PC S3 | 4 x 1 PC S3 |
|---|---|---|---|---|
| Expected length [bp] | 663 | 2,500 | 663 | 2,500 |
| Named | A -J (5 colonies per tube) | A -J (5 colonies per tube) | 1-4 | 1-4 |
| Primer fw 10 µM | 10 x 1 µl VF2 | 10 x 1 µl HM13 | 4 x 1 µl VF2 | 4 x 1 µl HM13 |
| Primer rev 10 µM | 10 x 1 µl DN07 | 10 x 1 µl VR2 | 1 µl DN07 | 1 µl VR2 |
| Dream-Taq Polymerase (2x) | 10 x 10 µl | 10 x 10 µl | 4 x 10 µl | 4 x 10 µl |
| ddH2O | 10 x 8 µl | 10 x 8 µl | 4 x 8 µl | 4 x 8 µl |
| Cycles | Temperature Screening start[°C] | Time [s] | Temperature Screening end [°C] | Time [s] |
|---|---|---|---|---|
| 1 | 95 | 120 | 95 | 120 |
| 12 | 95 | 60 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | 60 (touchdown -0.5°C) | 30 | |
| 72 | 45 | 72 | 2:30 min | |
| 12x/ 34x | 95 | 60 | 95 | 60 |
| 65 | 30 | 60 | 30 | |
| 72 | 45 | 72 | 2:30 min | |
| 1 | 72 | 5 min | 72 | 5 min |
| 1 | 12 | inf | 12 | inf |
Result
Expected band: 663 bp (screening start), 2.5 Kb (screening end)
Gel does not show expected bands.
- => Screen 10 minipreped colonies of the plate 5 and pick another 10 colonies of plate 1, 2, 3 and 6 (on plate 4 are no colonies).
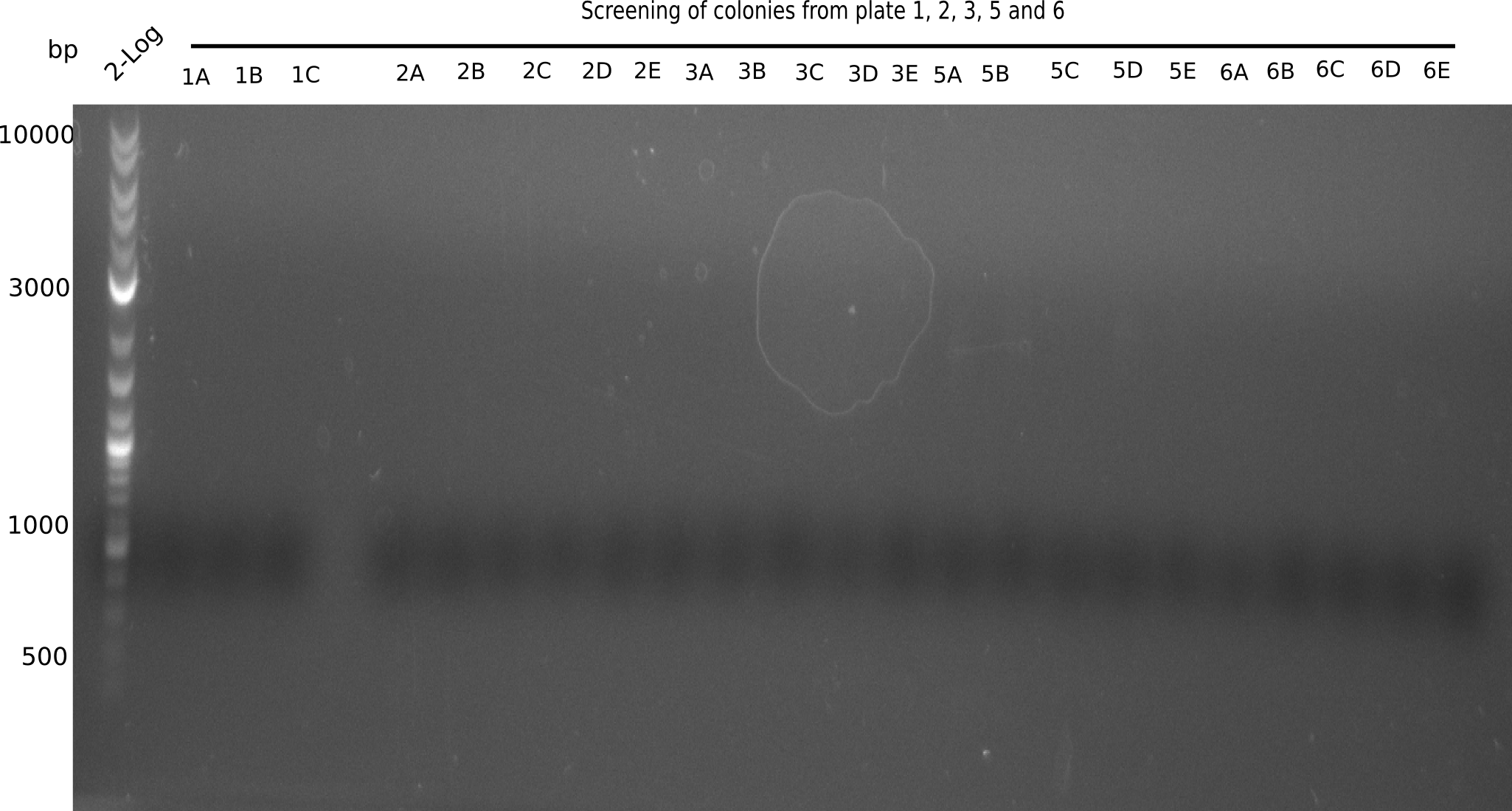
Colony-PCR CP.W17.D
- 2 different screening are performed
- PC = picked colony
- S5 = Sample 5 (watch names above on construct DelH-BB)
| Template | 10 x 1 PC S1 | 10 x 1 PC S1 | 10 x 1 PC S2 | 10 x 1 PC S2 | 10 x 1 PC S3 | 10 x 1 PC S3 | 10 x 1 PC S6 | 10 x 1 PC S6 | 10 x 1 µl of minipreped colony | 10 x 1 µl of minipreped colony |
|---|---|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 2.5 | 663 | 2.5 | 663 | 2.5 | 663 | 2.5 | 663 | 2.5 |
| Named | 1A-E (2 colonies per tube) | 1F-J (2 colonies per tube) | 2A-E (2 colonies per tube) | 2F-J (2 colonies per tube) | 3A-E (2 colonies per tube) | 3F-J (2 colonies per tube) | 6A-E (2 colonies per tube) | 6F-J (2 colonies per tube) | ............. | .................... |
| Primer fw 10 µM | 5 x 1 µl VF2 | 5 x 1 µl HM13 | 5 x 1 µl VF2 | 5 x 1 µl HM13 | 5 x 1 µl VF2 | 5 x 1 µl HM13 | 5 x 1 µl VF2 | 5 x 1 µl HM13 | 5 x 1 µl VF2 | 5 x 1 µl HM13 |
| Primer rev 10 µM | 5 x 1 µl DN07 | 5 x 1 µl VR2 | 5 µl DN07 | 5 µl VR2 | 5 x 1 µl DN07 | 5 x 1 µl VR2 | 5 µl DN07 | 5 µl VR2 | 5 x 1 µl DN07 | 5 x 1 µl VR2 |
| Dream-Taq Polymerase (2x) | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl | 5 x 10 µl |
| ddH2O | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 8 µl | 5 x 6 µl | 5 x 6 µl |
| Cycles | Temperature Screening start [°C] | Time [s] | Temperature Screening end [°C] | Time [s] |
|---|---|---|---|---|
| 1 | 95 | 120 | 95 | 120 |
| 12 | 95 | 60 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | 60 (touchdown -0.5°C) | 30 | |
| 72 | 45 | 72 | 2:30 min | |
| 12x/ 34x | 95 | 60 | 95 | 60 |
| 65 | 30 | 60 | 30 | |
| 72 | 45 | 72 | 2:30 min | |
| 1 | 72 | 5 min | 72 | 5 min |
| 1 | 12 | inf | 12 | inf |
Result
Expected band: 663 bp (screening start), 2.5 Kb (screening end)
None of the gels shows expected fragment.
- => Overthink strategy.
 "
"