|
|
| (444 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | | | |
| | ==Overview== | | ==Overview== |
| | + | <div align="justify"> |
| | | | |
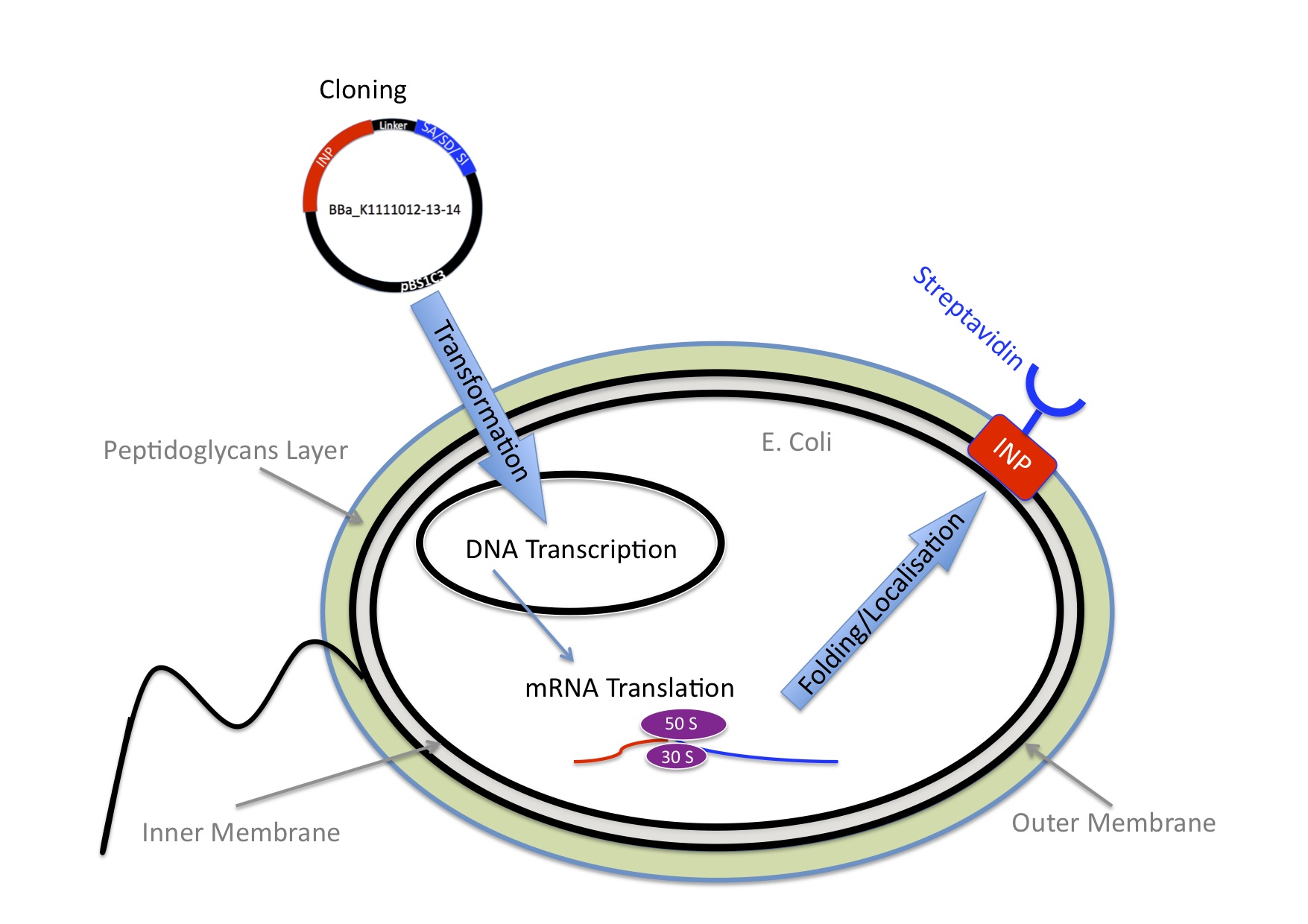
| - | ''How can one bind Nanoparticles to bacteria?''
| + | [[File:EPF-Lausanne-Overview-Coupling.jpg|thumb|right|350px|Figure 1:<br>Scheme of the cell surface display of streptavidin]] |
| | + | We knew we wanted to bind Nanoparticles to the Bacteria using the very strong interaction between biotin and streptavidin. |
| | + | <br> |
| | + | Next we needed to decide which strategy would be the best so the E.coli expresses streptavidin on the outer membrane. |
| | + | <br> |
| | + | E.Coli are Gram-negative bacteria and they have two membranes, an inner one and an outer one, separated by a periplasmic space. Furthermore, the outer membrane is covered with a glycocalix, a layer of sugars. |
| | + | <br> |
| | | | |
| - | [[File:EPF-Lausanne-Overview-Coupling.jpg|thumb|right|300px]]
| |
| | | | |
| - | Once we knew we wanted to bind Nanoparticles to bacteria we needed an idea how to connect those two components. We came up with the idea to use one of the strongest non covalent interactions existing in nature, the interaction between Streptavidin and Biotin with a Kd of 10-14 mol/l. Streptavidin is a tetrameric protein, each of the four subunits can bind one biotin. The Streptavidin-Biotin complex is resistant against different pHs, temperatures and detergents. Also streptavidin binds only rarely unspecific. The bacteria need to express Streptavidin on its surface and the Nanoparticles needed to be biotynilated.
| + | There are different ways to express a heterologous protein on the outer membrane of E.coli. Peptides can be fused with outer membrane proteins or on fimbriae, fused to beta-autotransporters or lipoproteins, as for example the (Lpp)-OmpA protein. But then there is also another promising display system based on the use of ice nucleation protein (INP) from Pseudomonas syringae. Which we decided to use. |
| | + | |
| | + | ==Our Biobricks== |
| | + | |
| | + | <html> |
| | + | <p align =center> |
| | + | <script> |
| | + | function hideinfos() { |
| | + | var infos = document.getElementsByClassName("biobrick")[0].getElementsByClassName("areaDescription") |
| | + | |
| | + | for(var i=0; i < infos.length; i++) |
| | + | { |
| | + | infos[i].style.visibility = "hidden"; |
| | + | } |
| | + | } |
| | + | |
| | + | function showinfo(infoid) { |
| | + | hideinfos(); |
| | + | document.getElementById(infoid).style.visibility = "visible"; |
| | + | } |
| | + | </script> |
| | + | |
| | + | <style type="text/css"> |
| | + | .biobrick { |
| | + | position:relative; |
| | + | display: block; |
| | + | margin-left: auto; |
| | + | margin-right: auto; |
| | + | width: 500px; |
| | + | } |
| | + | |
| | + | .biobrick > .areaDescription { |
| | + | position: absolute; |
| | + | width: 250px; |
| | + | border-bottom: 1px dotted #000000; color: #000000; outline: none; |
| | + | padding: 0.5em 0.8em 0.8em 2em; |
| | + | background: #ffee90; |
| | + | border: 1px solid #888; |
| | + | visibility: hidden; |
| | + | } |
| | + | |
| | + | #brickDescription { |
| | + | font-size: 94%; |
| | + | text-align: center; |
| | + | } |
| | + | </style> |
| | + | |
| | + | <div class="biobrick"> |
| | + | <img id="biobrickImg" src="https://static.igem.org/mediawiki/2013/thumb/4/46/Team-EPF-Lausanne_Biobrick.jpg/500px-Team-EPF-Lausanne_Biobrick.jpg" border="0" alt="Biobrick" usemap="#biobrickMap" onmouseover="hideinfos()"> |
| | + | <map id="biobrickMap" name="biobrickMap"> |
| | + | <area shape="poly" title="" coords="361,46,399,69,421,94,459,154,432,170,360,84,299,58,310,26" alt="" onmouseover="showinfo('areaBlau')"> |
| | + | <area shape="poly" title="" coords="191,30,197,59,135,84,72,161,51,238,52,259,16,261,18,196,51,116,121,49" alt="" onmouseover="showinfo('areaRot')"> |
| | + | <area shape="poly" title="" coords="199,31,242,20,293,26,293,52,244,48,197,54,196,27" alt="" onmouseover="showinfo('areaSchwarz')"> |
| | + | <area shape="poly" title="" coords="462,165,478,256,463,337,411,409,304,465,240,473,128,431,37,327,22,263,49,259,60,312,114,381,231,435,331,428,390,387,438,308,447,225,432,172" alt="" onmouseover="showinfo('areaSchwarz')"> |
| | + | </map> |
| | + | <p id="brickDescription">For Details, please move the cursor onto the image.</p> |
| | + | |
| | + | <p class="areaDescription" id="areaRot" style="left:40px;top:140px"><b>Ice nucleation Protein </b><br> INP originates from the bacteria Pseudomonas syringae. It is an outer membrane protein. That has successfully been used for cell surface display of enzymes, antibodies and antigens.</p> |
| | + | |
| | + | <p class="areaDescription" id="areaBlau" style="left:250px;top:100px"><b>Streptavidin</b><br>Streptavidin originates from the bacterium Streptomyces avidinii. It is a tetrameric protein, each of the four subunit can bind one biotin molecule with a very high affinity. |
| | <br> | | <br> |
| | + | For our Biobricks we used three different kinds of streptavidin. |
| | + | <br></p> |
| | + | |
| | + | <p class="areaDescription" id="areaSchwarz" style="left:120px;top:270px"><b>pBS1C3</b><br>The three biobricks were cloned into the standard iGEM backbone plasmid, which contains a chloramphenicol resistance for selection.</p> |
| | + | </div> |
| | + | </p> |
| | + | </html> |
| | + | |
| | + | We decided to engineer a fusion protein between Ice nucleation protein (INP) and Streptavidin. Because such fusion proteins have been successfully used for cell surface display of enzymes, single-chain antibodies and antigens. So we assumed it would work for streptavidin as well. Especially because Jung et al ([http://www.ncbi.nlm.nih.gov/pubmed/9624691 1]) even succeeded to express a functional surface enzyme, that was much bigger than streptavidin. |
| | <br> | | <br> |
| | + | Ice nucleation protein is a glycosyl-phosphatidyliositol anchored outer membrane protein, this motif is quite unique in prokaryotes. When it is expressed in E.coli over 90% of INP is found in the outer membrane, Wolber et al ([http://www.pnas.org/content/83/19/7256.short 2]). |
| | + | |
| | + | '''Streptavidin'''<br> |
| | + | Streptavidin originates from the bacterium Streptomyces avidinii ([http://www.ncbi.nlm.nih.gov/nuccore/A00743.1 NCBI]). It is a tetrameric protein, and each of the four subunits can bind biotin with a very high affinity. |
| | + | |
| | + | ''We cloned three Biobricks that contain different versions of streptavidin.'' |
| | + | <br\> |
| | + | |
| | + | '''BBa_K1111012''' |
| | <br> | | <br> |
| | + | Encodes a fusion protein between INP and a monovalent streptavidin provided by the laboratory of Mark Howarth at Oxford university Department of Biochemistry ([http://users.ox.ac.uk/~bioc0756/MyWebs/activesite/index.htm 3]) |
| | <br> | | <br> |
| | + | '''BBa_K1111013''' |
| | <br> | | <br> |
| | + | Encodes a fusion protein between INP and a dead version of streptavidin. It was designed to serve as a negative control for the microscopy functional assays, because its affinity to biotin is much lower. |
| | <br> | | <br> |
| | + | '''BBa_K1111014''' |
| | + | <br> |
| | + | Encodes a fusion protein between INP and streptavidin we received in the Biobrick BBa_K283010 ([http://parts.igem.org/Part:BBa_K283010 BBa_K283012, Main Page]). |
| | + | <br><br> |
| | | | |
| - | ==INP-Streptavidin== | + | ==INP-YFP Characterization== |
| - | E.Coli are Gram-negative bacteria and they have two membranes, an inner one and an outer one, separated by a periplasmic space. Furthermore, the outer membrane is covered with a glycocalix, a layer of sugars. (add image)
| + | |
| - | There are different strategies to express a heterologous protein on the outer membrane of E.coli. Peptides can be inserted in outer membrane proteins, or on fimbriae, fused to beta-autotransporters or lipoproteins, as for example the (Lpp)-OmpA protein5 which was used by the igem team 2006 of Harvard and the igem team 2009 of Washington. But then there is also another promising display system that uses ice nucleation protein (INP) from pseusomonas syringae, which is a glycosyl-phosphatidyliositol anchored outer membrane protein. The GPI anchor is normally used by eukaryotes, so to see this motif in prokaryotes is quite unique.7 Over 90% of INP is found in the outer membrane when it is expressed in E.coli (Wolber et al). It has successfully been used for cell surface display of enzymes, single-chain antibodies and antigens.9
| + | |
| - | Jung et al (1998) successfully expressed functional levansucrase on the surface of E.coli by fusing it to INP, levansucrase is a 47 kDA protein. Streptavidin is only 14 kDa, so we assumed that it should be even easier for the E.coli to export it.
| + | |
| | | | |
| - | Streptavidin originates from the bacterium Streptomyces avidinii. It is a tetrameric protein, which means it has 4 subunits, each of them can bind a biotin molecule. For our Biobricks we used different kinds of streptavidin.
| + | There is a Biobrick that contains INP in the iGEM registry (BBa_K523013). It was designed by the [https://2011.igem.org/Team:Edinburgh/Cell_Display Edinburgh 2011 iGEM team]. This Biobrick encodes a fusion protein of a truncated and codon optimized INP and yellow fluorescent protein (YFP) under control of the Lac promoter. The INP is truncated, it only contains the first 211 and last 97 amino acids that are normally encoded by the inaK gene. This is enough to target it to the outer membrane. |
| - | The Biobrick BBa_K1111014 contains streptavidin that was cloned out of the Biobrick BBa_K283010. | + | |
| | | | |
| - | The Biobrick BBa_K1111012 has a monomeric streptavidin in it, which means only one of the four subunits of streptavidin is able to bind to biotin. This decreases the dissociation rate. The plasmid containing the sequence was provided by the laboratory of Mark Howarth at Oxford university Dept. of Biochemistry (http://users.ox.ac.uk/~bioc0756/MyWebs/activesite/index.htm).
| + | <br> |
| - | The Biobrick BBa_K1111013 was designed to serve as a negative control for our experiments, it contains a dead version of streptavidin, which means it doesn’t bind to biotin.
| + | We improved the Biobrick <html> <a href= "http://parts.igem.org/Part:BBa_K523013">BBa_523013</a> </html> by extensively characterizing it. |
| | + | <br> |
| | | | |
| - | ==Our Biobricks==
| |
| - | [[File:Team-EPF-Lausanne Biobrick.jpg|center|500px]]
| |
| - | onmouseover="This is our biobrick()"
| |
| | | | |
| - | ==INP-YFP Characterization==
| + | We used confocal microscopy to see if it was expressed and eventually show surface localization. |
| | + | <br> |
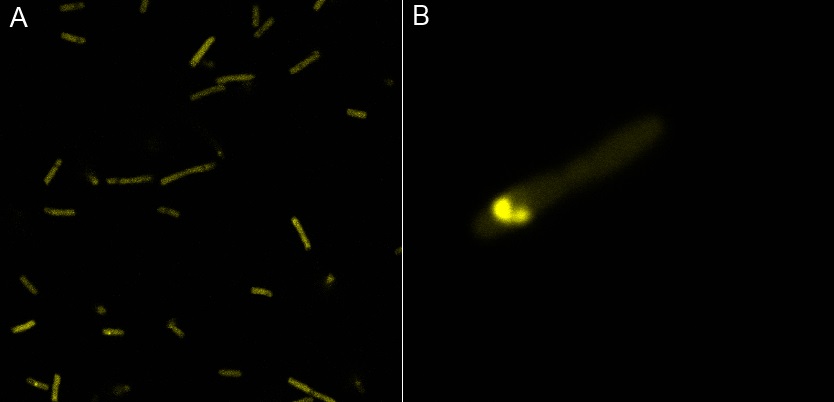
| | + | Thanks to Thierry Laroche for taking the images with the confocal microscope. |
| | + | <!--Confocal Microscopy--> |
| | + | [[File:Team-EPF-Lausanne_INP-WFPconfocal.jpg|thumb|700px|left|Figure 2:<br>'''Confocal image of DH5alpha E.coli transformed with BBa_K523013'''<br>A: shows YFP fluorescence (514 nm)<br> B: zoom of one Bacteria that shows spot of clustered YFP ]] |
| | | | |
| - | There was already a Biobrick that contained INP in the registry (BBa_K523013), so we decided to use it. It was designed by the Edinburgh igem team of 2011 (https://2011.igem.org/Team:Edinburgh/Cell_Display). This Biobrick encodes a fusion protein of a truncated and codon optimized INP and yellow fluorescent protein (YFP) under control of the LacZ promoter. The INP is truncated, it only contains the first 211 and last 97 amino acids that are normally encoded by the inaK gene.
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| | + | |
| | + | Looking at E.coli we noticed that fluorescence was localized in specific specks, that the team of Edinburgh didn’t describe. We started looking for an explanation for this phenomena and found in the literature that INP forms aggregates in the cell membranes [http://www.ncbi.nlm.nih.gov/pubmed/2203606 [3]] This suggests that INP also forms aggregates when it is fused to other proteins. We continued characterizing this biobrick by first determining the percentage of bacteria that were fluorescent. We found that approximately 30% of E.coli expressed YFP. |
| | + | <br><br> |
| | + | |
| | + | The team of Edinburgh grew bacteria and lysed them. After centrifugation, they looked where the fluorescence seemed concentrated. On the transformed bacteria, it was mainly the pellet that appeared fluorescent, meaning that the fusion protein was embedded in the membrane. The negative control, in contrast, showed fluorescence in the pellet as well as in the supernatant with the same intensity. This method was used because their microscopy was not good enough. |
| | <br> | | <br> |
| - | We improved the Biobrick BBa_K523013 by extensively characterizing it.
| + | The confocal microscope was really powerful. But we too, didn't succeed to proof surface localization of the fusion protein by simply looking at the intrinsic fluorescence of the YFP, because the signal was to diffuse. |
| | + | <br><br> |
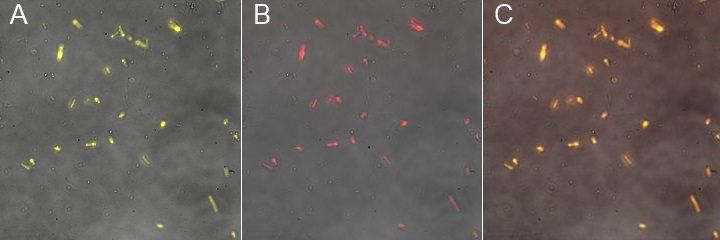
| | + | So we came up with an alternative strategy: '''Immunofluorescent assay''' using an anti-YFP antibody and visualizing it with an inverted microscope. |
| | + | <br> |
| | + | [[File:Team-EPF-Lausanne_INP-WFP-merged-3pics.jpg|thumb|700px|left| Figure 3:<br>'''Inverted microscope composite images of BBa_K523013 transformed DH5alpha E.coli'''<br> A: shows YFP fluorescence (514 nm)<br> B: stained with biotinylated anti-YFP antibody and avidin dylight (650 nm)<br> C: Merge of A and B, shows colocalization of fluorescent signal]] |
| | | | |
| - | We used confocal microscopy (Thanks to Thierry Laroche for operating the microscope) to see if it was expressed and eventually show surface localization.
| |
| - | <!--Confocal Microscopy-->
| |
| - | [[File:Team-EPF-Lausanne INP-YFP-confocal.jpg|thumb|250px|left|]]
| |
| - | [[File:EPFL-INP-YFP-zoom.png|thumb|259px|left]]
| |
| | | | |
| | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| | | | |
| - | Looking at the image we realized that there were some points that showed very strong fluorescence , that the team of Edinburgh didn’t describe. We continued characterizing this biobrick by first determing the percentage of bacteria that was fluorescent, approximatively 30% of E.coli expressed YFP. We also counted the percentage of fluorescent bacteria that showed these points of very strong fluorescence, around XX%. We started looking for an explanation for this phenomena and found in the literature1, 2 that INP forms aggregates in the cell membrane, to have good ice nucleation abilities. So it suggests, that this also happens if INP is fused to another protein.
| + | We did an immunofluorescent assay with an anti YFP antibody (Thanks Francesca Volpetti for giving us the antibody). In Figure 3 you can see the results we obtained with microscopy. The YFP signal (Figure 3 A) and the Cy5 signal (The antibody was tagged with NeutrAvidin DylLight Fluorescent Conjugate red, 650 nm from pierce) (Figure 3 B) superimpose nicely (Figure 3 C). Thus proofing that the fusion protein of INP and YFP is exported to the outer membrane. |
| | + | <br><br><br> |
| | | | |
| - | The team of Edinburgh used an image of eppendorf tube to proof surface localization, because their imaging technology wasn’t good enough. We too didn't succed to proof surface localization by simply looking at the bacteria with a microscope. So we came up with an alternative strategy an immunofluorescent assay with an anti GFP antibody (which also binds to YFP) (Thanks to XXX for providing us with the antibody) . Then we made images with the microscope first a bright field, then exiting the YFP, and finally exiting the Antibody (red) By superimposing the images we could show nicely the two different fluorescent signals are colocalized. This proofs conclusively that the fusion protein between INP and YFP is exported to the outer membrane.
| + | ==Results== |
| - | [[File:Team-EPF-Lausanne_INP-YFP-AB-composite.jpg|thumb|center|500px|Merge image of the YFP signal and the anti-YFP-Antibody signal]]
| + | |
| | | | |
| - | <!--Images Reults-->
| + | In the following section, you will find the results concerning our streptavidin cell surface display work. |
| - | [[File:Team-EPF-Lausanne INP-YFP%.jpg|thumb|225px|left]]
| + | |
| - | [[File:Team-EPF-Lausanne INP-YFP-AB-R-G.jpg|thumb|225px|left]]
| + | |
| - | [[File:Team-EPF-Lausanne INP-YFP-AB-R-Y.jpg|thumb|225px|left]]
| + | |
| - | <br><br><br><br><br><br><br><br><br><br>
| + | |
| | | | |
| - | [[File:Team-EPF-Lausanne beads-1-biotin-BF-G.jpg|thumb|225px|left]]
| + | <br>Our first experimental steps were to perform Polymerase Chain Reactions (PCR) in order to isolate the different streptavidins. |
| - | [[File:Team-EPF-Lausanne Merged CC IPTG biotin.tif|thumb|225px|left]]
| + | |
| - | [[File:Team-EPF-Lausanne Merged ISA2 IPTG biotin5.jpg|thumb|225px|left]]
| + | |
| - | <br><br><br><br><br><br><br><br><br><br>
| + | |
| | | | |
| - | [[File:Team-EPF-Lausanne Merged ISA2 IPTG biotin4.jpg|thumb|250px|left]]
| + | <br>'''''Cloning''''' |
| - | [[File:Team-EPF-Lausanne Merged ISA2 IPTG biotin2.jpg|thumb|250px|left]]
| + | |
| | | | |
| - | <br><br><br><br><br><br><br><br><br><br> | + | <!--PCR images--> |
| | + | |
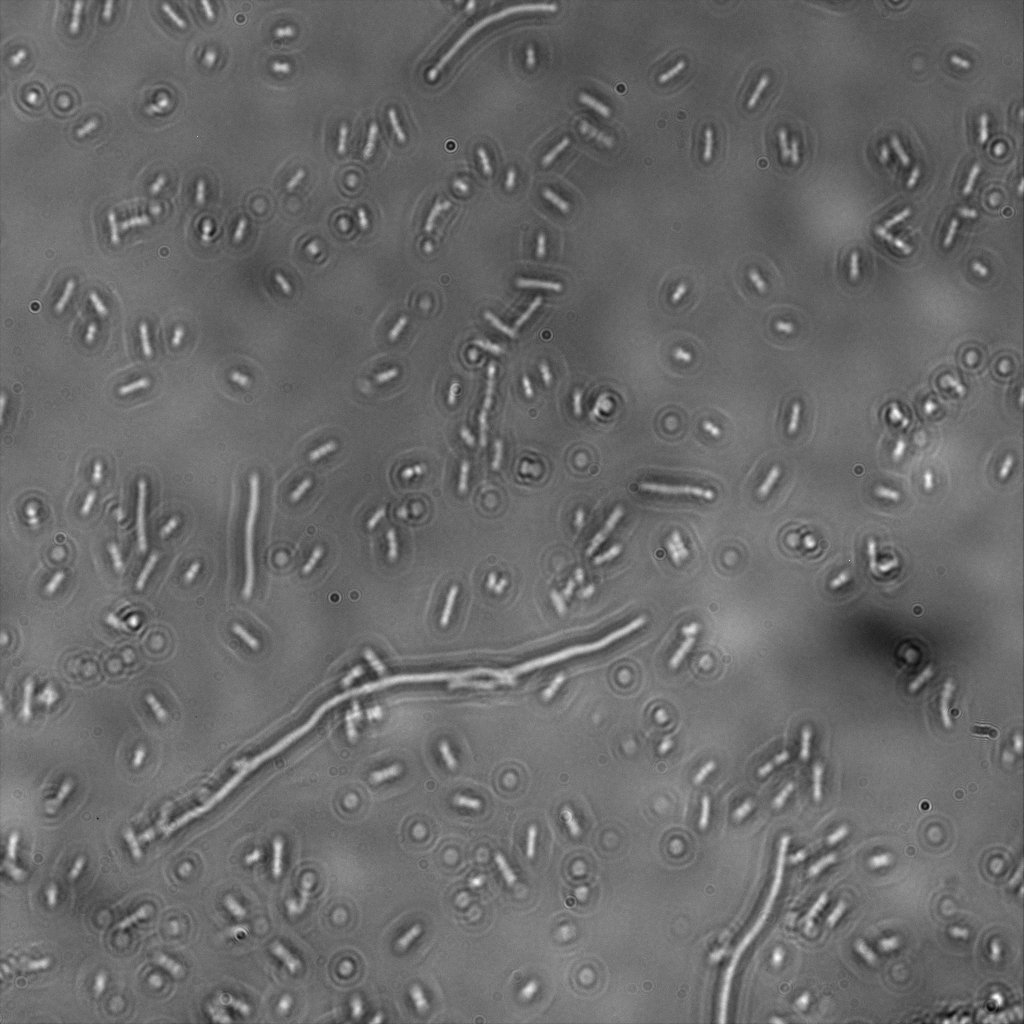
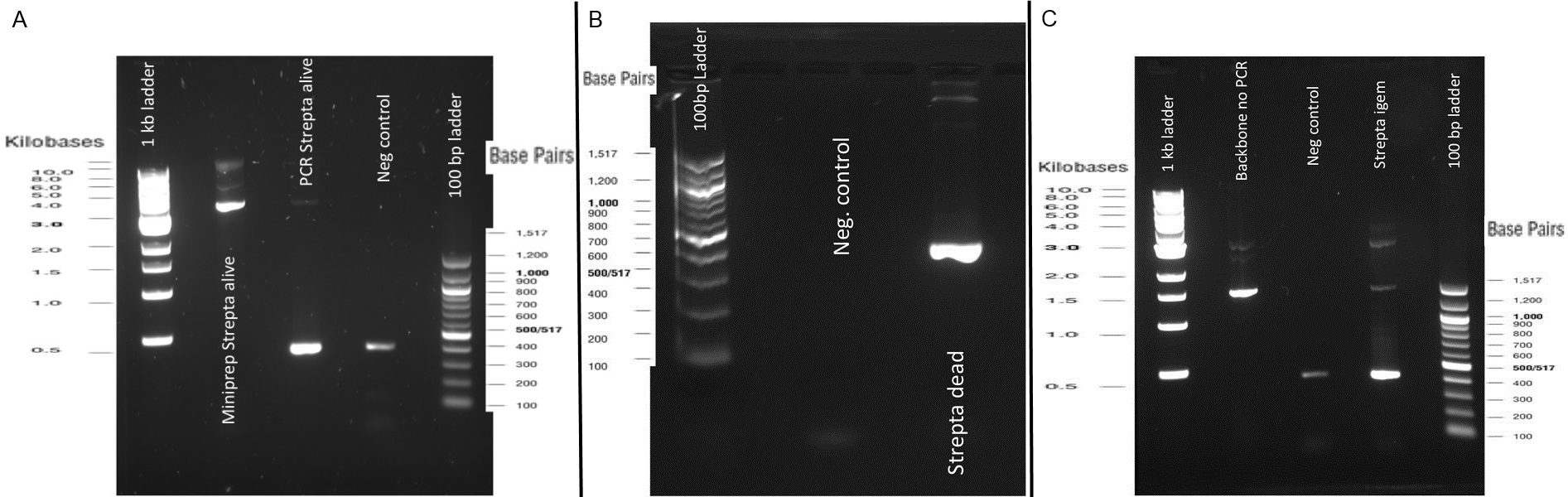
| | + | [[File:Team-EPF-Lausanne_ISX.jpg|thumb|800px|center|Gel A: 0.8% Gel electrophoresis to verify PCR of Streptavidin Alive coding sequence. Expected fragment size is 384bp and nothing in the reagent negative control. |
| | + | <br>Gel B: 0.8% Gel electrophoresis to verify PCR of Streptavidin Dead coding sequence. Expected fragment size is 387bp and nothing in the reagent negative control |
| | + | <br>Gel C: 0.8% Gel electrophoresis to verify PCR of Streptavidin iGEM coding sequence. Expected fragment size is 387bp and nothing in the reagent negative control]] |
| | + | <br> |
| | + | The previous Gels show that our PCR reactions (68°C annealing temp, primers in registry) to amplify the streptavidin coding sequences went fine. The DNA fragments migrated at the expected sizes written under each images. The PCR to amplify our backbone (see primers in the registry) from the BBa_K523013 biobrick and to add overhangs complementary to the edges of the streptavidin sequences also worked. |
| | + | <br><br> |
| | + | With the PCR products we did a Gibson Assembly to obtained our desired constructs. |
| | + | The Gibson Assembly reaction worked as expected, we obtained a product around the expected size 4171bp. |
| | + | <br> |
| | + | <br>Then another way to verify that our Gibson assembly reaction had worked was to transform cells with the Gibson products. The transformed DH5α competent cells should be able to grow on an antibiotic medium because of the resistance provided in the plasmid. The only problem with this second verification is that the Gibson product could still contain parental plasmid, whose insertion in cells can also lead to cells resistance and survival. So to verify if there is a carry over of the initial plasmids, we made negative controls. |
| | + | <br>You can see bellow plates containing cells from the glycerol stocks we made of our gibson products . |
| | + | |
| | + | <!--Plates images--> |
| | + | [[File:Team-EPF-Lausanne_plate_ISA.jpeg|thumb|240px|left|Image 1: INP_Streptavidin Alive Gibson transformed cells plated from the glycerol stock ]] |
| | + | [[File: Team-EPF-Lausanne_plate_ISD.jpeg|thumb|240px|left|Image 2: INP_Streptavidin Dead Gibson transformed cells plated from the glycerol stock]] |
| | + | [[File: Team-EPF-Lausanne_plate_ISI.jpeg|thumb|240px|left|Image 3: INP_Streptavidin iGEM Gibson transformed cells plated from the glycerol stock]] |
| | + | <br><br><br><br><br><br><br><br><br><br><br><br> |
| | + | |
| | + | As the other experiments did not provide information about possible mutations, we also sent our constructs for sequencing, using VR and VF2 iGEM primers but also another forward primer we designed ourselves. The sequencing results were good, the protein is well inserted and there is no frame shift. You can see the sequencing results using the following iGEM registry links: <html><a href="http://parts.igem.org/Part:BBa_K1111012:Experience">results_ISA </a>; <a href="http://parts.igem.org/Part:BBa_K1111013:Experience">results_ISD </a>;<a href="http://parts.igem.org/Part:BBa_K1111014:Experience">results_ISI </a></html>. |
| | + | |
| | + | So up to now, with the previous information we had on our constructs, we could simply conclude that our basic cloning strategy worked. |
| | + | |
| | + | |
| | + | Then moving on, we needed to test if the protein was expressed, if it was well localized at the outer membrane and that it was functional. |
| | + | We thus designed immunostaining assays to verify localization at the outer membrane and biotin-staining assay for the protein functionality ([https://2013.igem.org/Team:EPF_Lausanne/Protocols see protocols]). |
| | + | Then we visualized the samples using a NIKON Eclipse TI inverted microscope [http://en.wikipedia.org/wiki/Inverted_microscope] and also a Confocal microscope [http://en.wikipedia.org/wiki/Confocal_microscopy], which is one of the most powerful microscopes nowadays. |
| | + | Unfortunately for all these assays, we didn't have an appropriate positive control as our constructs were newly designed by ourselves. |
| | + | <br><br> |
| | + | |
| | + | '''''Confocal Microscopy''''' |
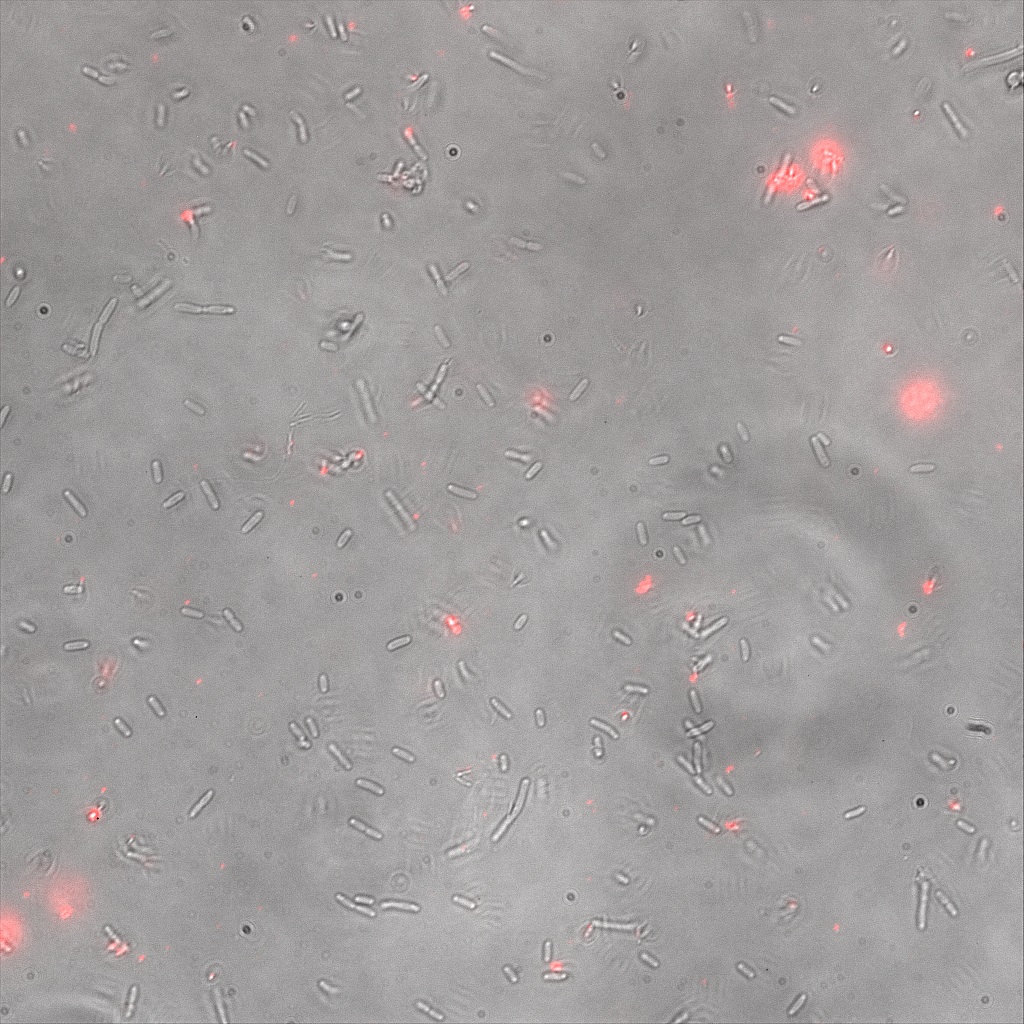
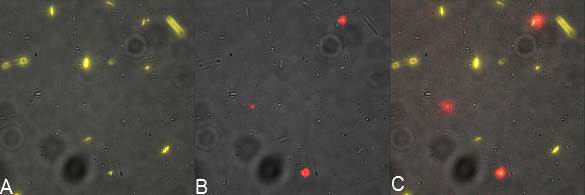
| | + | <br>Focusing on the biotin assays, we did the confocal experiment to better characterize this observed phenomena. You can observe the results below (Figure 1 to 4). |
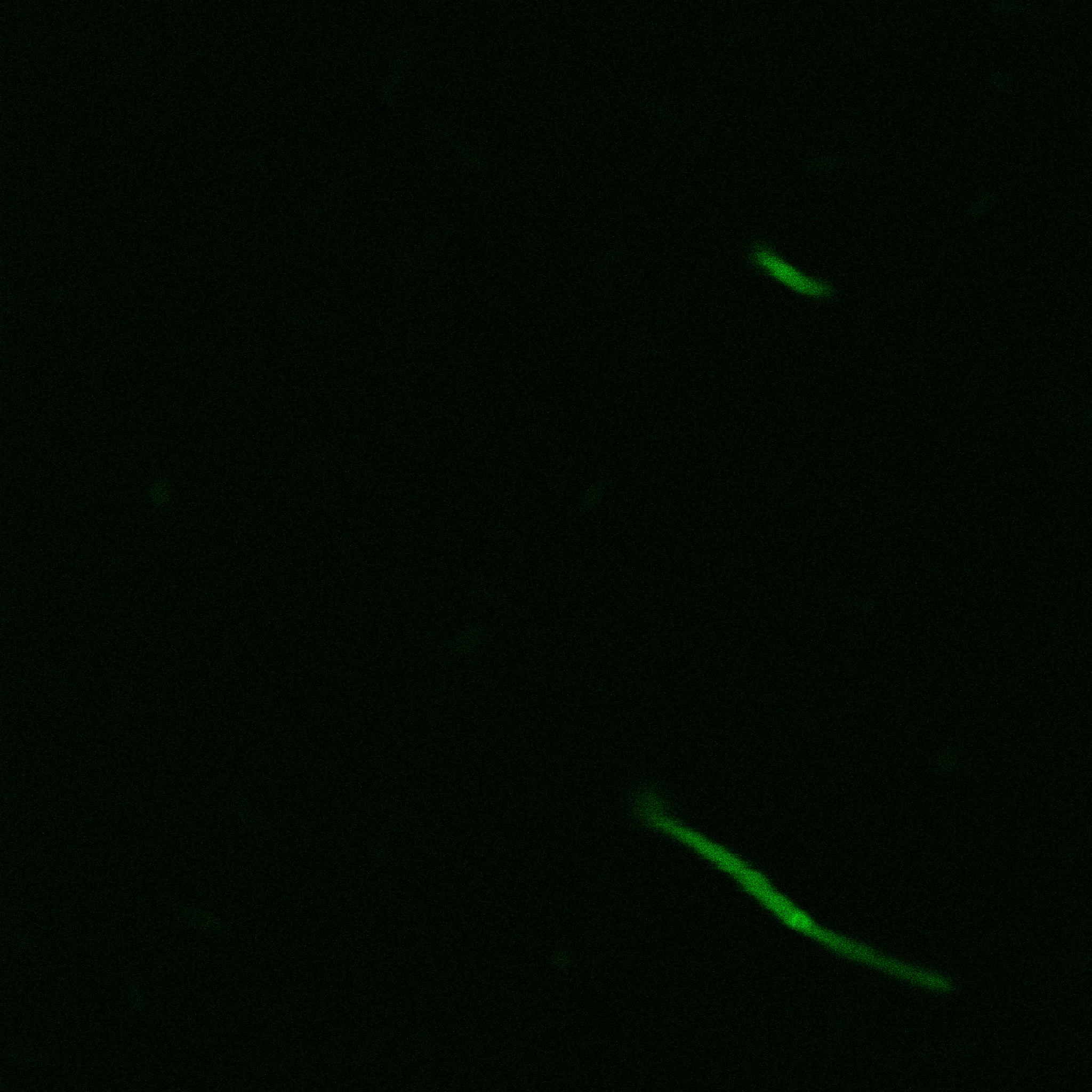
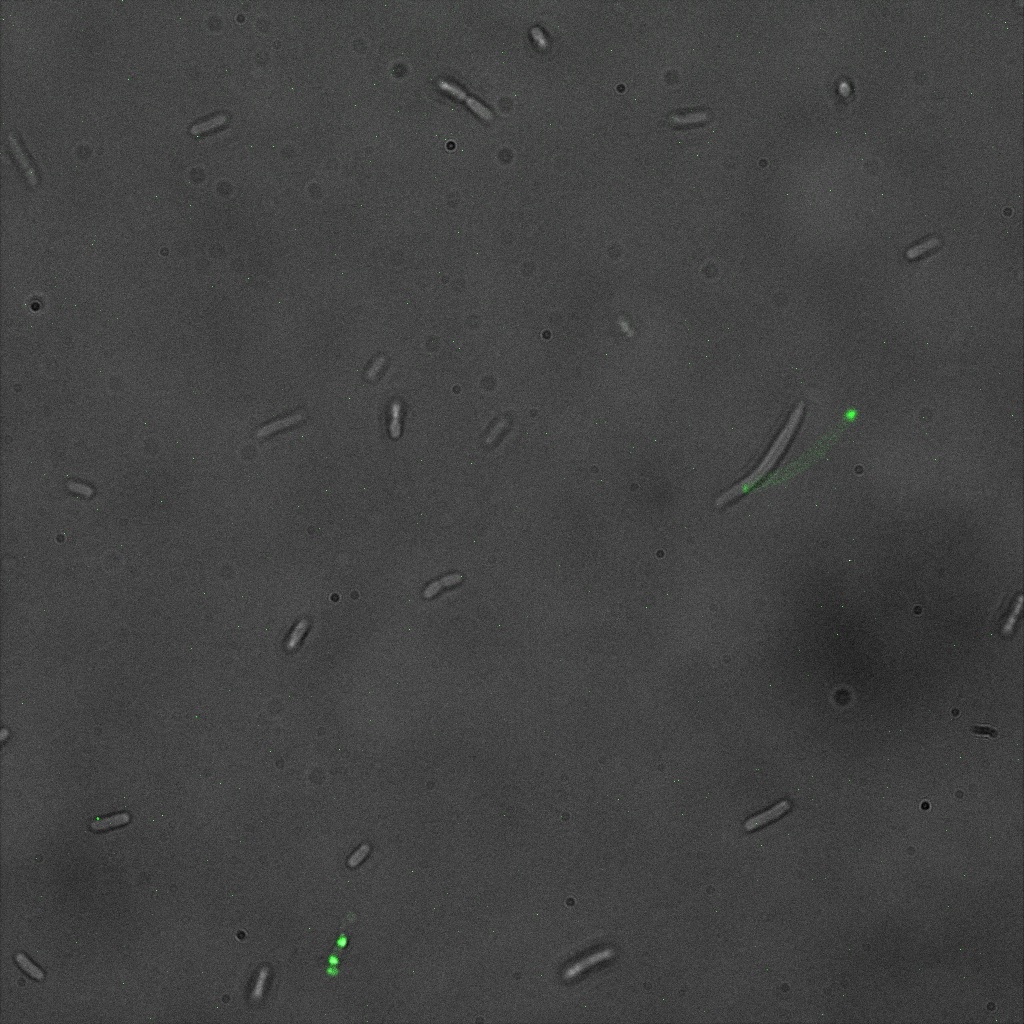
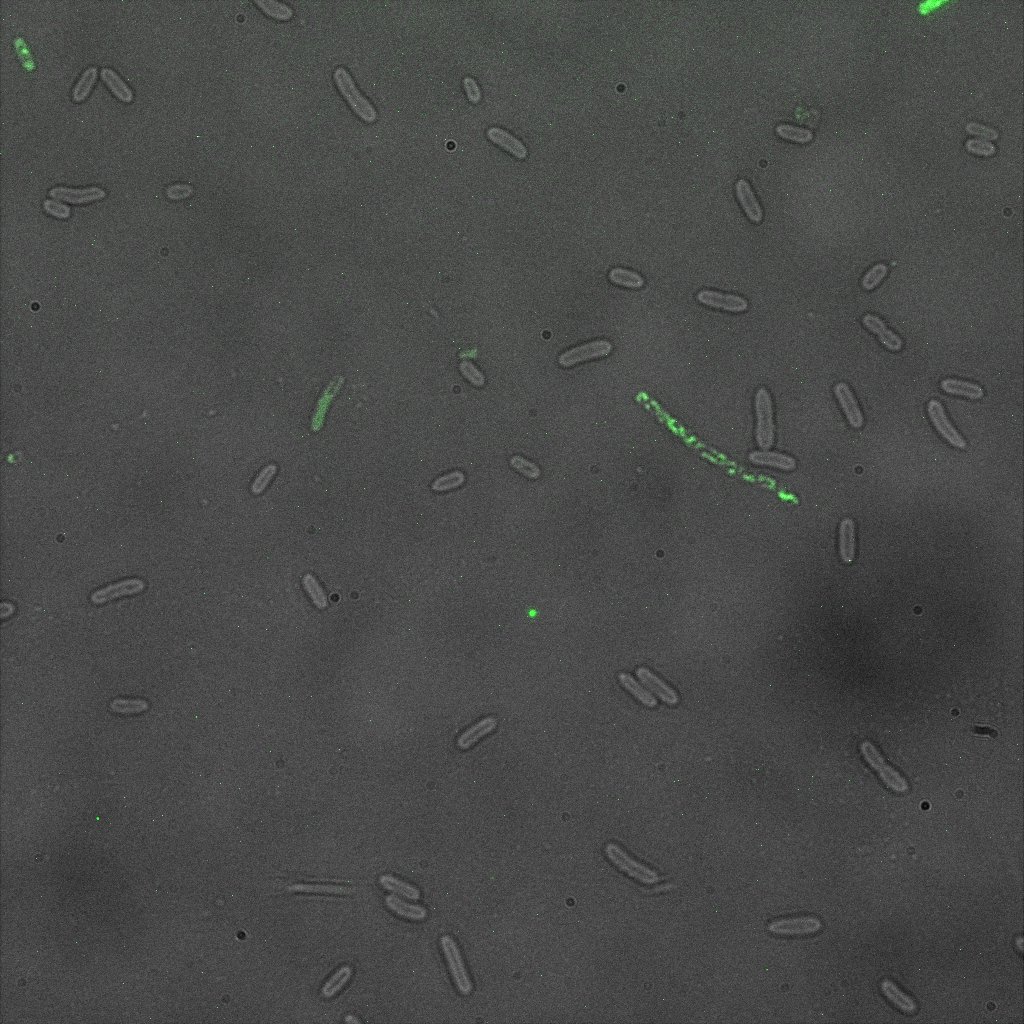
| | + | [[File:Team-EPF-Lausanne_ISA_biotin_conf.jpg|thumb|350px|left|Figure 1: confocal microscopy image showing ISA cells incubated 1hr with biotin conjugated to FITC reporter]] |
| | + | [[File: Team-EPF-Lausanne_ISD_biotin_conf.jpg|thumb|350px|right|Figure 2: confocal microscopy image showing ISD cells incubated 1hr with biotin conjugated to FITC reporter]] |
| | + | <br><br> |
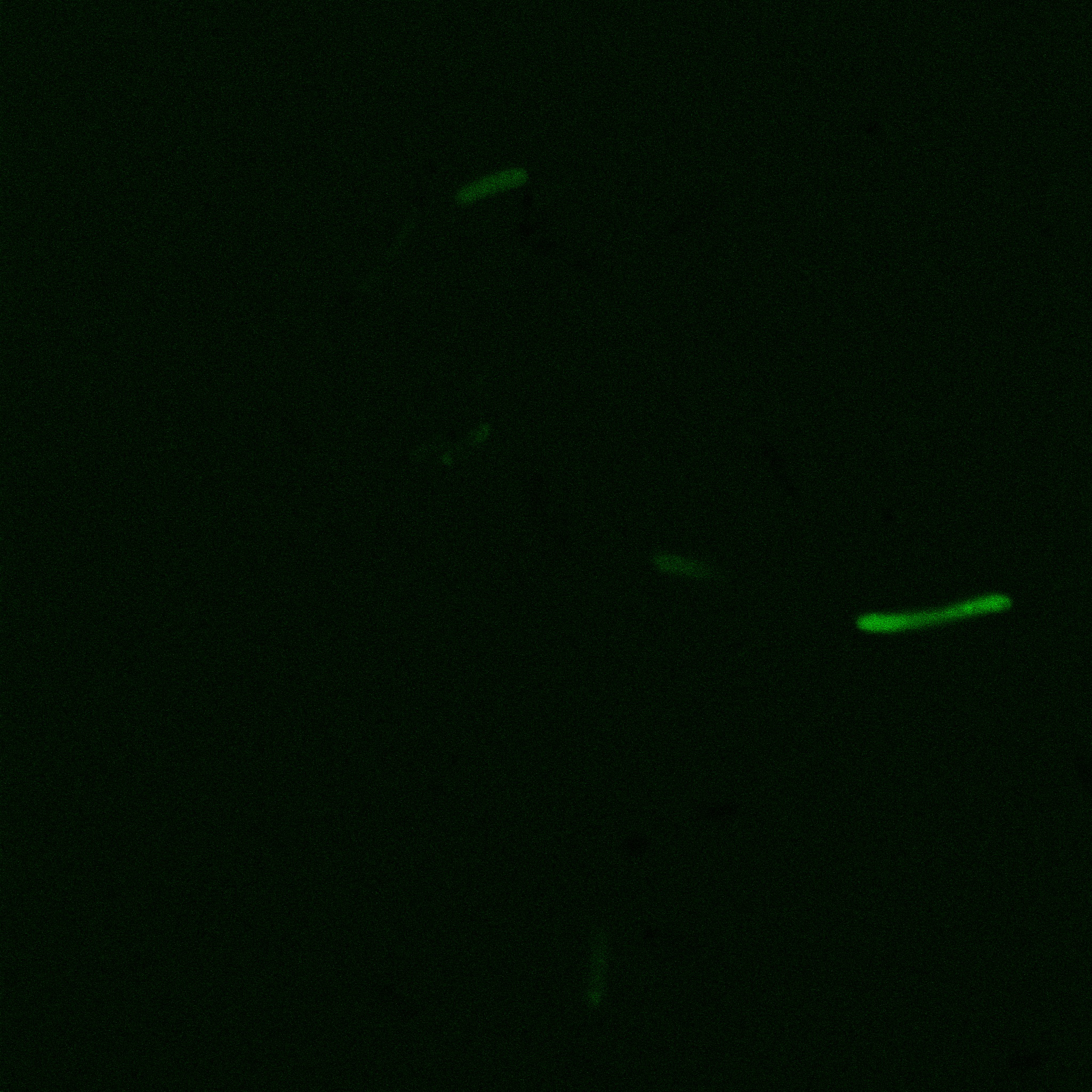
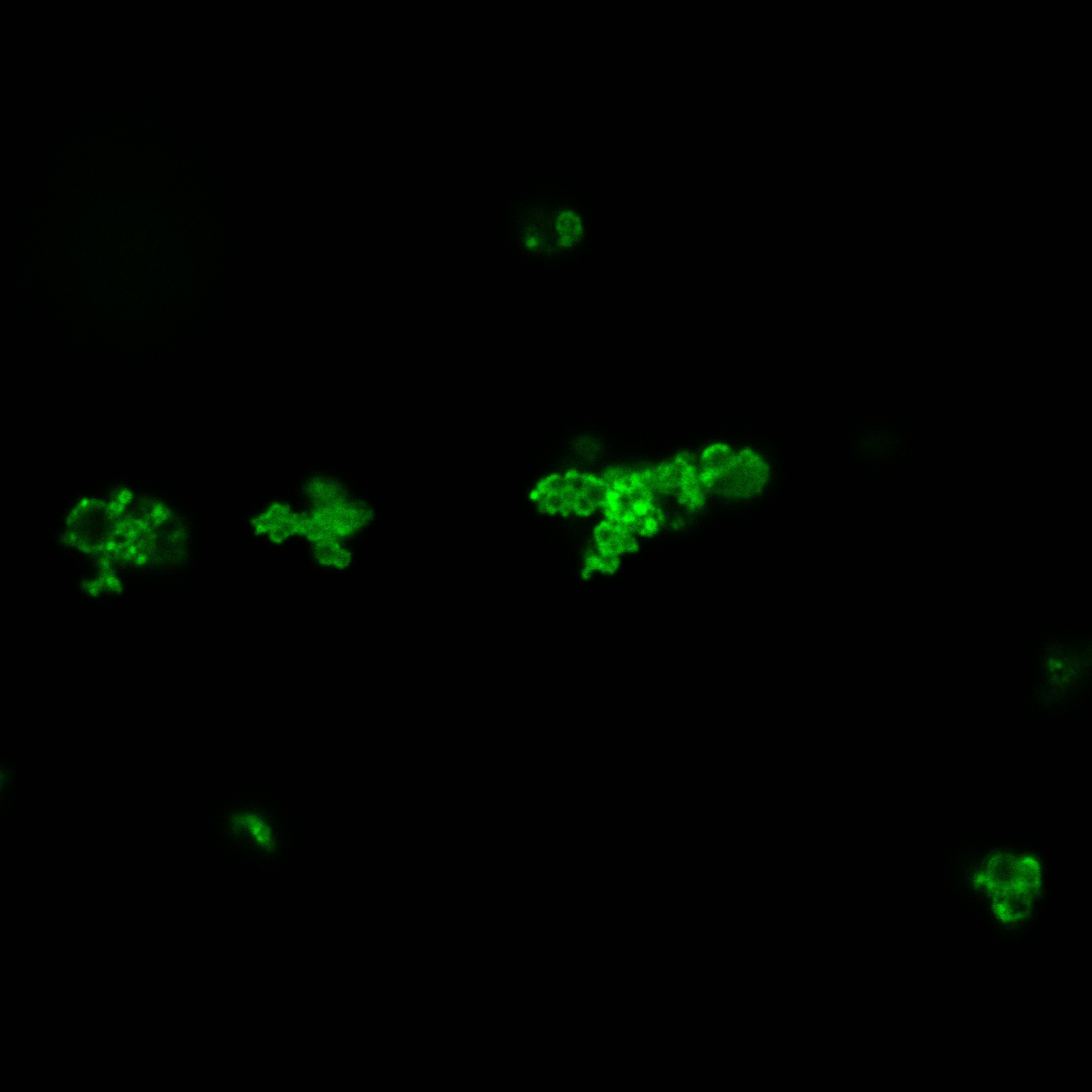
| | + | [[File: Team-EPF-Lausanne_CC_biotin_conf.jpg|thumb|350px|left|Figure 3: confocal microscopy image showing Competent cells incubated 1hr with biotin conjugated to FITC reporter]] |
| | + | [[File:Team-EPF-Lausanne_bead_biotin_conf.jpg|thumb|350px|right|Figure 4: confocal microscopy image showing streptavidin coated magnetic beads incubated 1hr with biotin conjugated to FITC reporter]] |
| | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| | + | |
| | + | With the confocal microscopy, we were not able to distinguish weather the biotin molecules were inside or on the outside of the cells. And also we can see that some cells in the negative control are fluorescent. But these results were promising so we decided to repeat the experiments, trying to optimize the biotin staining protocols we were using. |
| | + | |
| | + | <!--microscopy images--> |
| | + | |
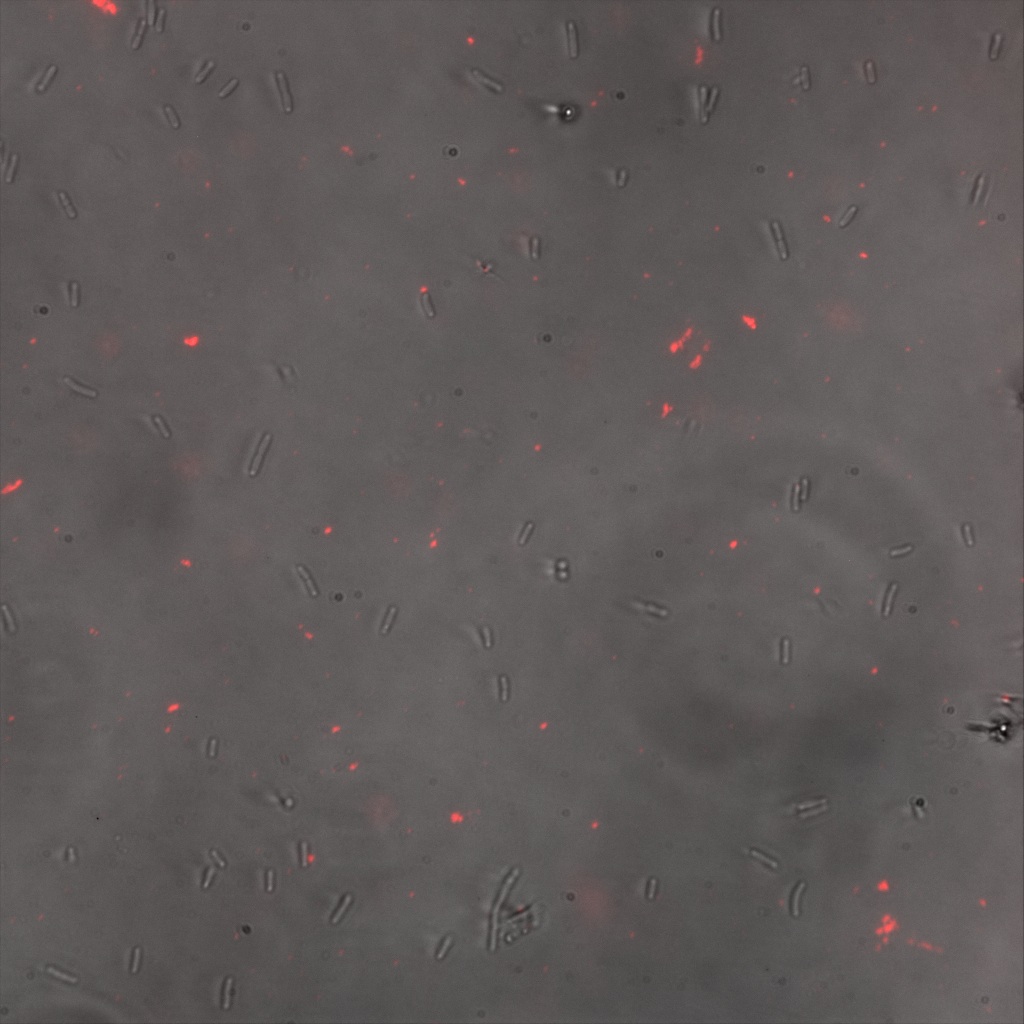
| | + | '''''Inverted Microscopy''''' |
| | + | |
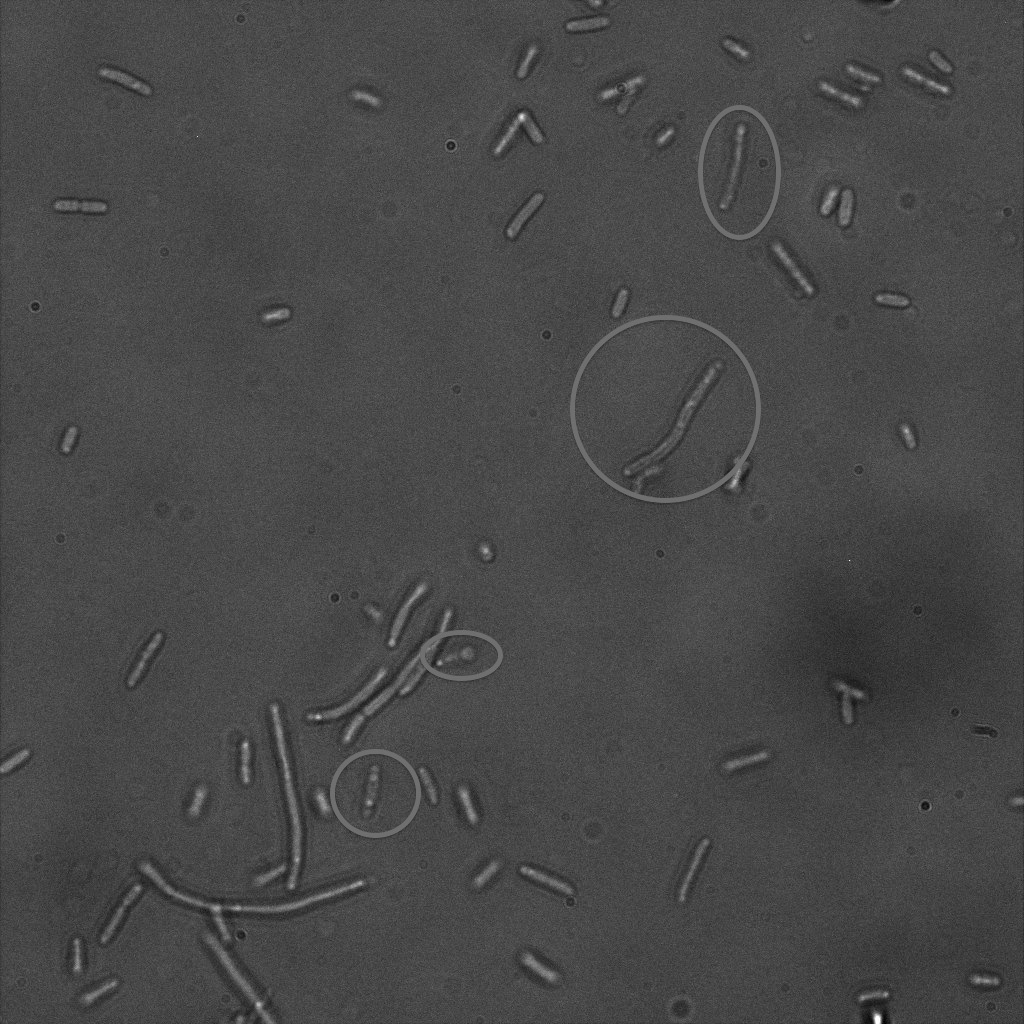
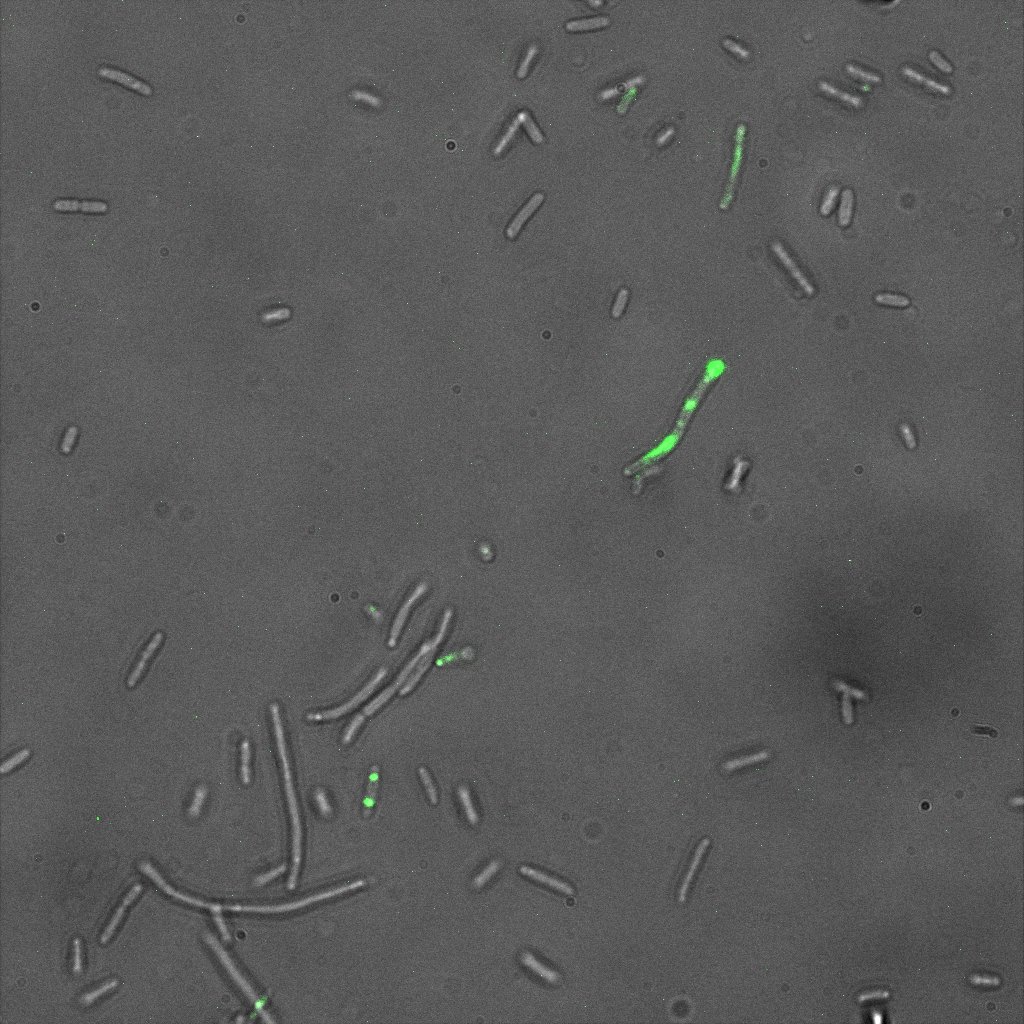
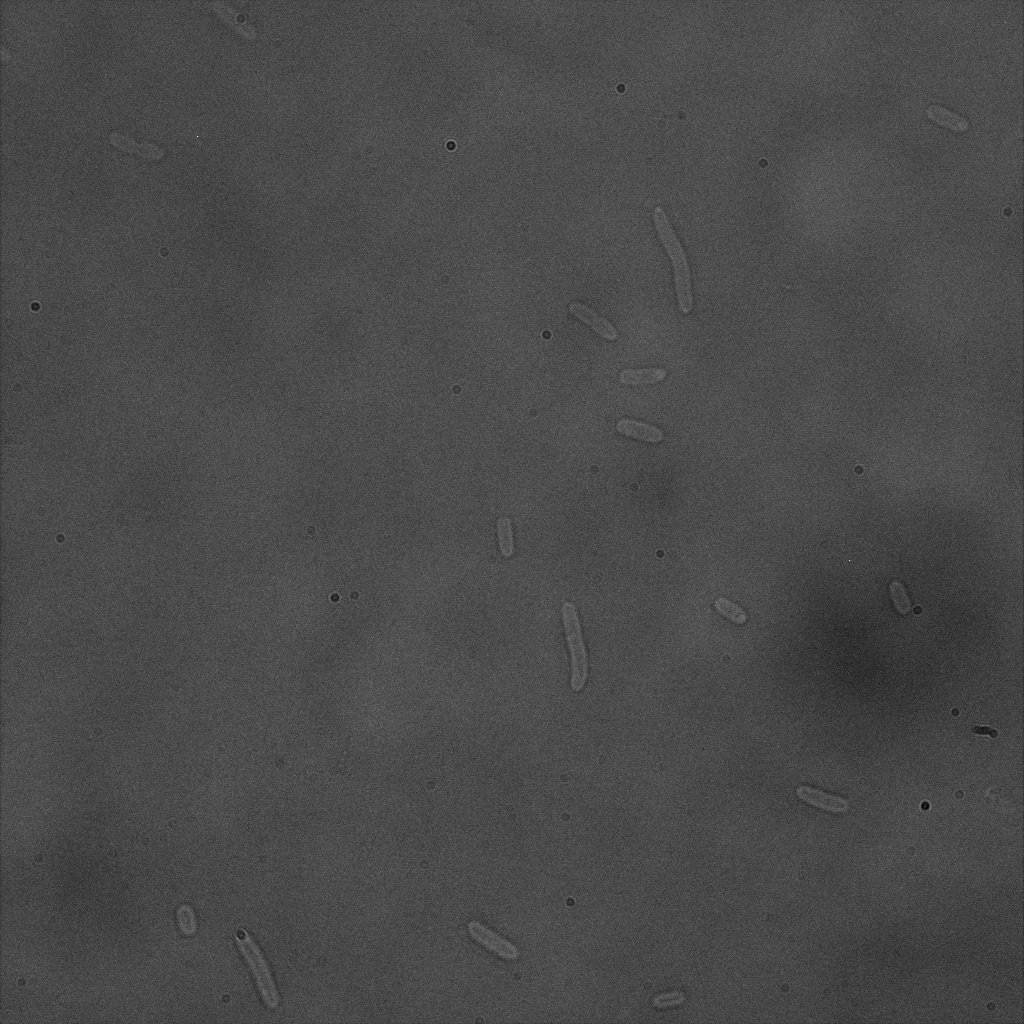
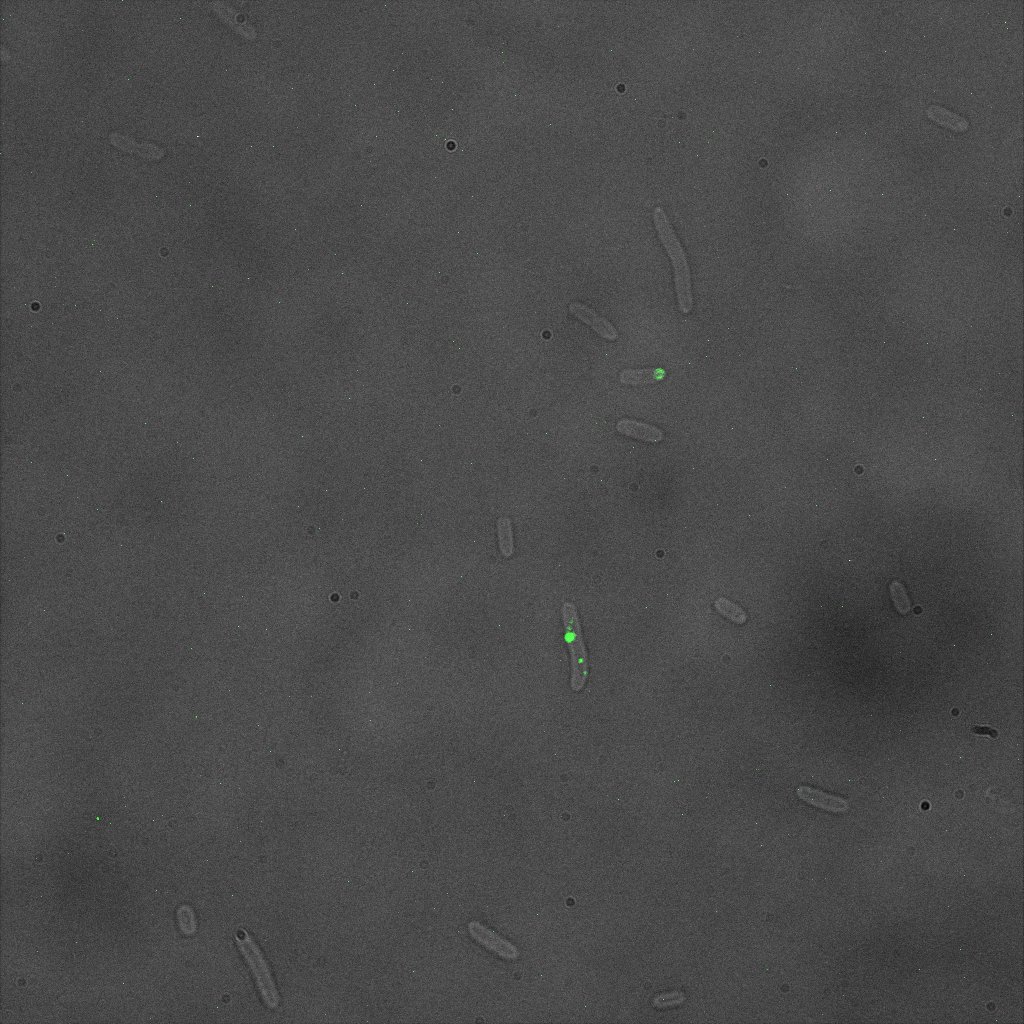
| | + | The four following images represent only the construct ISA. You can observe the bright field images followed by their corresponding composite images (bright field file merged with fluorescent file). |
| | + | <br> |
| | + | [[File:Team-EPF-Lausanne_ISA_biotin_bf.jpg|thumb|350px|left|Figure 5.1: Bright field image of ISA cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter ]] |
| | + | [[File: Team-EPF-Lausanne_ISA_biotin_RGB.jpg|thumb|350px|right|Figure 5.2: Composite image of ISA cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | <br><br> |
| | + | [[File: Team-EPF-Lausanne_ISA5_bf.jpg|thumb|350px|left|Figure 6.1: Bright field image of ISA cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | [[File:Team-EPF-Lausanne_ISA5_biotin_3.10(RGB).jpg|thumb|350px|right|Figure 6.2: Composite image of ISA cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | |
| | + | <!--<br><br> |
| | + | [[File:Team-EPF-Lausanne_ISA3_bf.jpg|thumb|177px|left|Figure 7.1: Bright field image of ISA cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter ]] |
| | + | [[File: Team-EPF-Lausanne_ISA3_bition_RGB.jpg|thumb|177px|left|Figure 7.2: Composite image of ISA cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | [[File: Team-EPF-Lausanne_ISA4_bf.jpg|thumb|177px|left|Figure 8.1: Bright field image of ISA cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | [[File:Team-EPF-Lausanne_ISA4_biotin_RGB.jpg|thumb|177px|left|Figure 8.2: Composite image of ISA cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]]--> |
| | + | |
| | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| | + | |
| | + | You can observe localized specks on the cells surface. As expected, this pattern corresponds to the one we had while characterizing the INP_YFP fusion protein. |
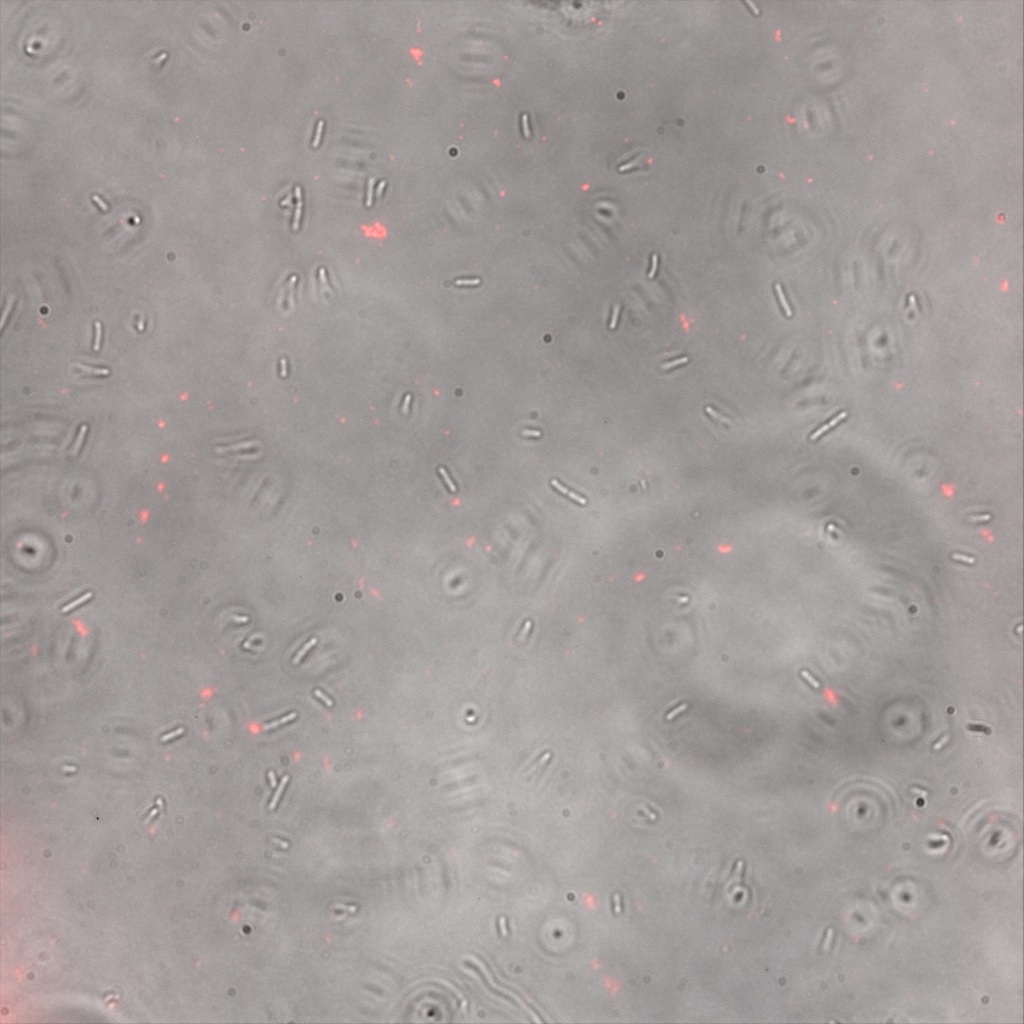
| | + | <br>The next images show the competent cells we use as negative controls, in the same conditions as explained above. |
| | + | |
| | + | <br> |
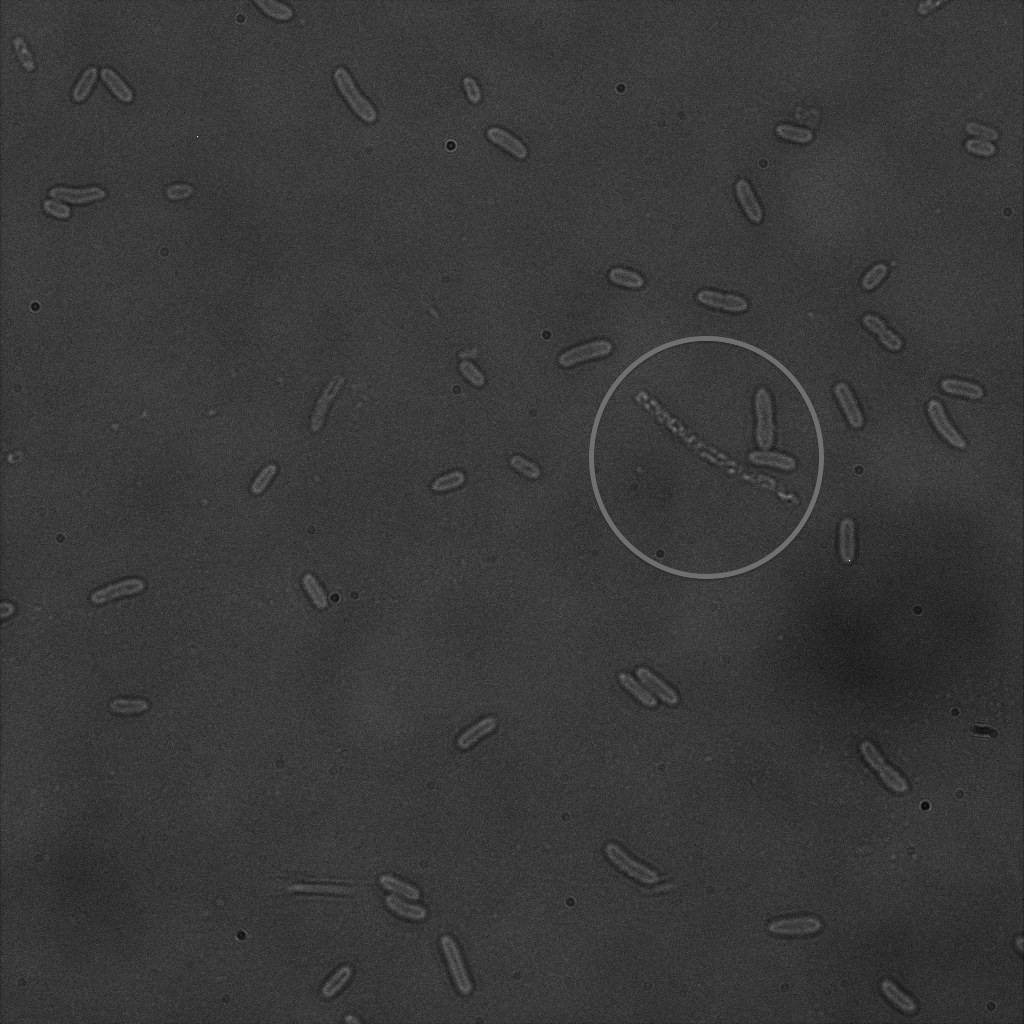
| | + | [[File:Team-EPF-Lausanne_CC5_bf.jpg|thumb|350px|left|Figure 7.1: Bright field image of competent cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter ]] |
| | + | [[File: Team-EPF-Lausanne_CC5_(RGB).jpg|thumb|350px|right|Figure 7.2: Composite image of competent cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| | + | <!--[[File: Team-EPF-Lausanne_CC7_bf.jpg|thumb|177px|left|Figure 8.1: Bright field image of competent cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | [[File:Team-EPF-Lausanne_CC7_RGB.jpg|thumb|177px|left|Figure 8.2: Composite image of competent cells after overnight incubation with 1mM IPTG and 1hr incubation with biotin conjugated to FITC reporter]]--> |
| | + | |
| | + | <br> It was not expected to find a pattern similar to the one found in the cells transformed with our constructs. |
| | + | <br>The next images corresponds to the two other constructs ISD and ISI. The cells look exactly like the ISA cells of Figures 1 and 2. |
| | + | |
| | + | <br><br> |
| | + | [[File:Team-EPF-Lausanne_ISD_bf.jpg|thumb|350px|left|Figure 9.1: Bright field image of ISD cells after 1hr incubation with biotin conjugated to FITC reporter ]] |
| | + | [[File: Team-EPF-Lausanne_ISD_biotin_RGB.jpg|thumb|350px|lright|Figure 9.2: Composite image of ISD cells 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | [[File: Team-EPF-Lausanne_ISI_bf.jpg|thumb|350px|left|Figure 10.1: Bright field image of ISI cells after 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | [[File:Team-EPF-Lausanne_ISI_biotin_RGB.jpg|thumb|350px|right|Figure 10.2: Composite image of ISI cells after 1hr incubation with biotin conjugated to FITC reporter]] |
| | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| | + | |
| | + | We thus decided to incubate ISA, ISD, ISI cells and competent cells with the some biotinylated nanoparticles conjugated with TexRed reporter. |
| | + | <br>This time again you can observe in the following pictures, nanoparticles (red) and cells, but we cannot tell if they are attached or not. |
| | + | |
| | + | [[File:Team-EPF-Lausanne_Isa_NP_RGB.jpg|thumb|350px|left|Figure 11: Composite image of ISA cells 1hr incubation with biotin coated nanoparticules conjugated to TexRed reporter]] |
| | + | [[File: Team-EPF-Lausanne_Isd_NP_RGB.jpg|thumb|350px|right|Figure 12: Composite image of ISD cells 1hr incubation with biotin coated nanoparticules conjugated to TexRed reporter]] |
| | + | [[File: Team-EPF-Lausanne_Isi_NP_RGB.jpg|thumb|350px|left|Figure 13: Composite image of ISI cells 1hr incubation with biotin coated nanoparticules conjugated to TexRed reporter]] |
| | + | [[File:Team-EPF-Lausanne_CC_NP_RGB.jpg|thumb|350px|right|Figure 14: Composite image of competent cells 1hr incubation with biotin coated nanoparticules conjugated to TexRed reporter]] |
| | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| | + | |
| | + | With the previous results we cannot make a conclusion on the functionality and localization of our fusion proteins. |
| | + | <br> We also did immunostaining assays, consisting in incubating the cells with FITC fluorescent antibody but we didn't have a fluorescent signal. |
| | + | With all this even if we know that our cloning strategy was good, we cannot really conclude anything about the protein's expression and functionality. |
| | + | |
| | + | <br><br><br> |
| | + | |
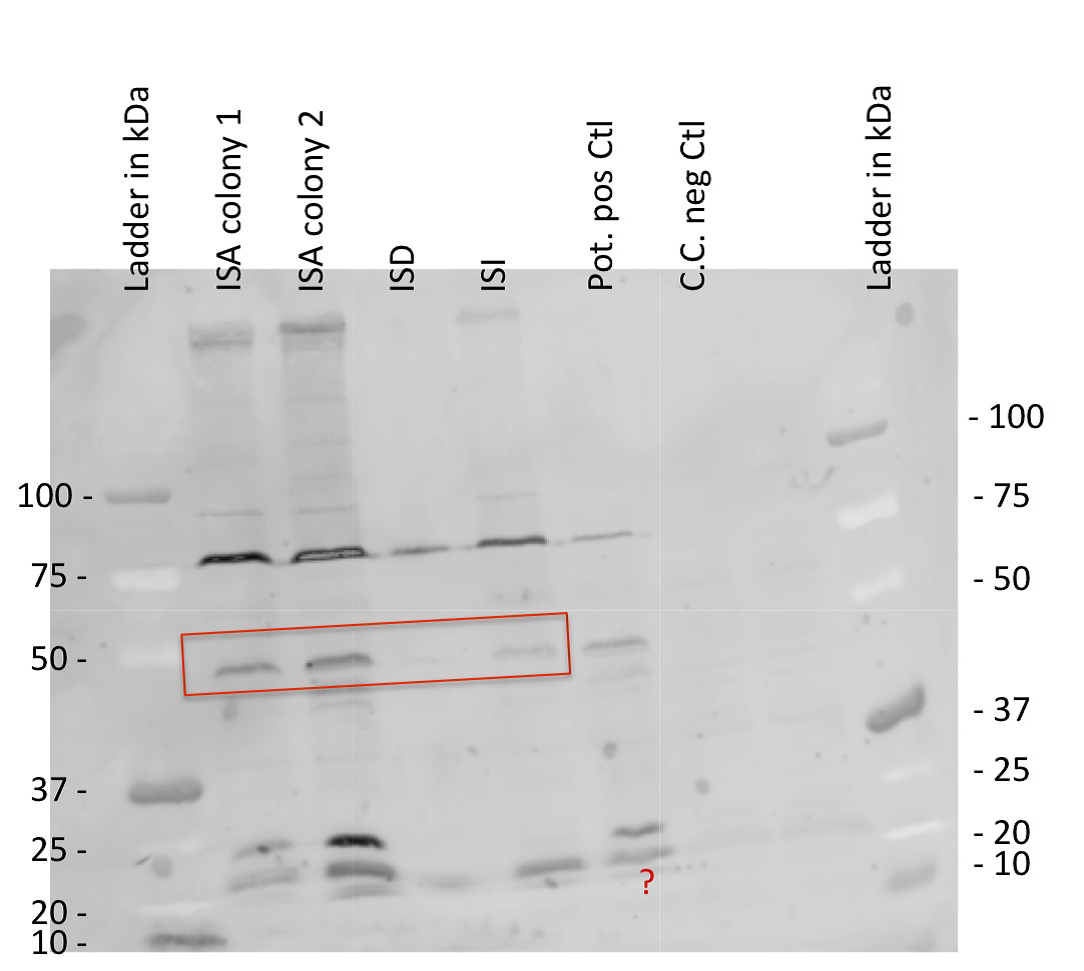
| | + | '''''Growth curve and Western Blot''''' |
| | + | <br> Finally we did a Western Blot and growth curve of our transformed cells to at least prove that our protein is indeed expressed. |
| | + | [[File:EPF-Lausanne_WB.jpg|thumb|left|327px|Figure15 : WB of the different INP - streptavidin constructs]] |
| | + | We did a western blot expecting a signal at the ~50kDa bands corresponding to our protein. For this experiment we used a FITC conjugated, anti-streptavidin polyclonal Antibody. Despite the gel quality, you can see in the following picture a band of the desired size. |
| | + | |
| | + | [[File:Team-EPF-Lausanne_Gcurve.jpg|border|425px|right]] |
| | + | |
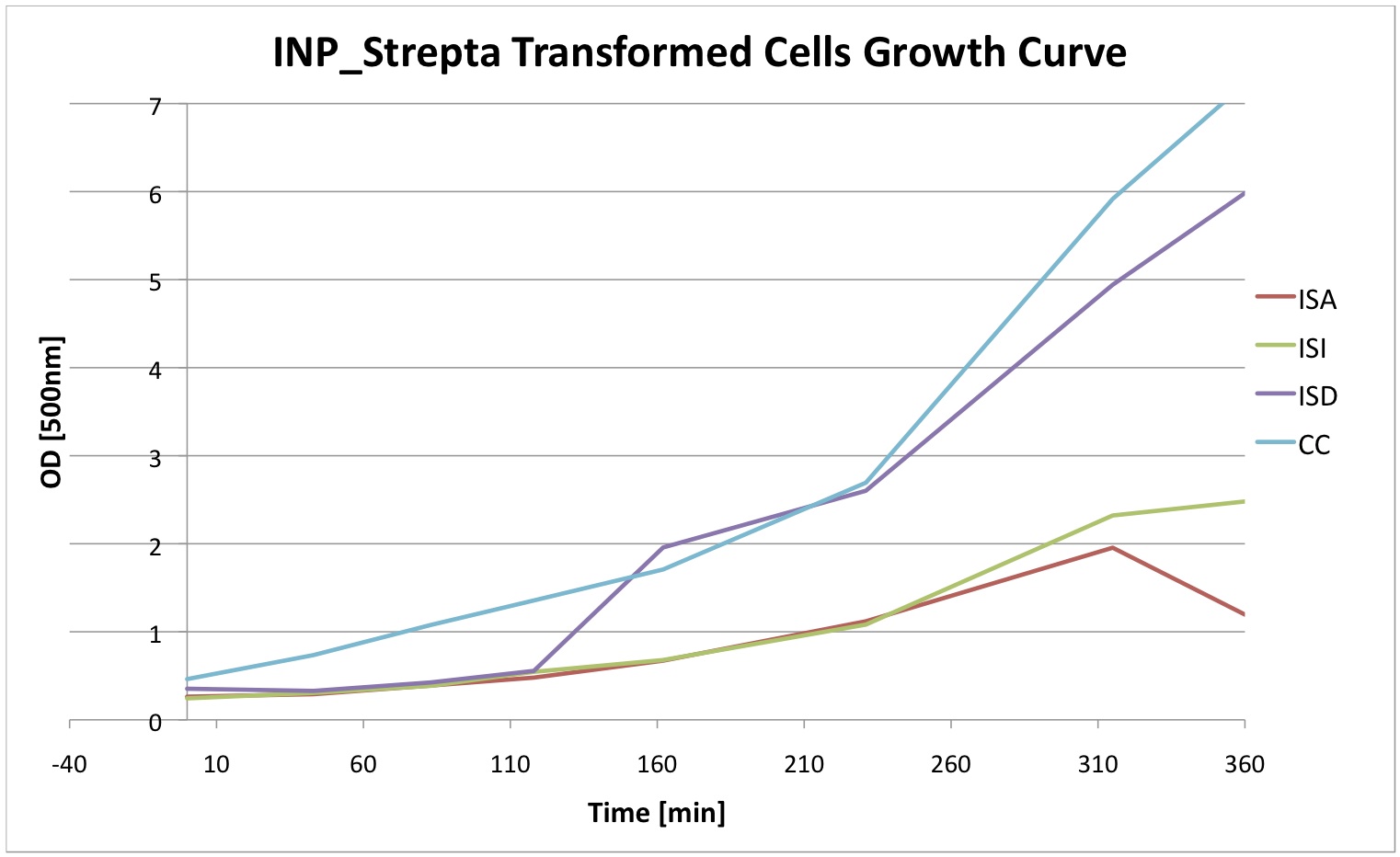
| | + | Finally we did a growth curve of the cells containing our constructs. For this last experiment, the cells were inoculated overnight with 1mM IPTG. In the morning they were initially diluted to a certain OD and then the OD was regularly measured. We can see that the insertion of our plasmid affects the growth of E. coli cells. |
| | + | <br><br> |
| | + | |
| | + | ==Discussion== |
| | + | |
| | + | <br> Our idea was to clone a fusion protein made of INP and Streptavidin that will be able to target streptavidin at the cell surface. This streptavidin could then further be used to attached biotinylated nanoparticules to the cells. We thus designed assays to clone this fusion protein and verify its functionality and localization. For the biotin assay, we were expecting ISA, ISI cells to bind biotin and ISD, competent cells to not bind it. We had the right patterns for ISA and ISI. We thus though that we had things working, but by repeating experiments several times and looking carefully at the competent and the ISD cells we found fluorescent cells also having the same characteristics as the ISA and ISI cells, which is unfortunately a contradictory result. We also had our immunostaining experiments that didn’t work, but the western blot results were positive. |
| | + | So knowing that we did all this very carefully with a strict scientific approach we came to the conclusion that our protein is expressed but might be localized inside the cells. It could be that it is not well folded preventing the fusion product to behave exactly like BBa_K523013 protein. |
| | + | <br><br> |
| | + | |
| | + | ==New Biobricks/Microscopy after the Jamboree in Lyon== |
| | + | |
| | + | With the additional time we designed new Biobricks <html><a href= "http://parts.igem.org/Part:BBa_K1111015">BBa_K1111015</a> & <a href= "http://parts.igem.org/Part:BBa_K1111016">BBa_K1111016</a></html>. |
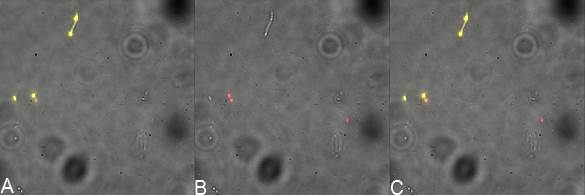
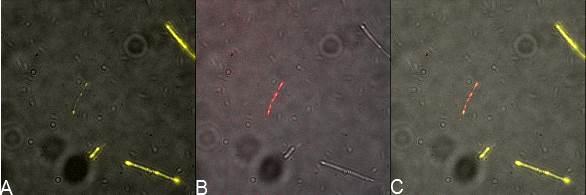
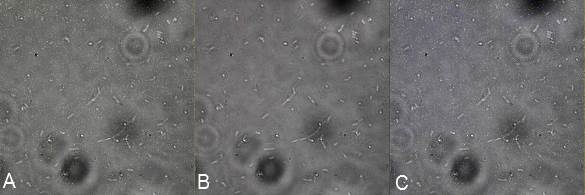
| | + | in order to prove surface localization of streptavidin. We designed a plasmid that contained INP-Streptavidin-YFP and one plasmid that encodes INP-YFP-Streptavidin. Indeed YFP should act as a reporter is the protein is expressed and could be target by an antibody as we did to characterize BBa_K523013. In addition an anti-Streptavidin antibody could be used to see colocalistion of the signals thus showing that the streptavidin is also at the outer membrane. In the following you see the results of the microscopy. |
| | + | |
| | + | [[File:Team-EPF-Lausanne_ISY.jpg|thumb|900px|centre| Figure X:<br>'''Inverted microscope composite images of INP-Streptavidin-YFP construct transformed DH5alpha E.coli'''<br> A: shows YFP fluorescence (514 nm)<br> B: stained with biotinylated anti-YFP antibody and avidin dylight (650 nm)<br> C: Merge of A and B, shows colocalization of fluorescent signal]] |
| | + | [[File:Team-EPF-Lausanne_IYS.jpg|thumb|800px|centre| Figure X:<br>'''Inverted microscope composite images of INP-YFP-Streptavidin construct transformed DH5alpha E.coli'''<br> A: shows YFP fluorescence (514 nm)<br> B: stained with biotinylated anti-YFP antibody and avidin dylight (650 nm)<br> C: Merge of A and B, shows colocalization of fluorescent signal]] |
| | + | [[File:Team-EPF-Lausanne_NEGCTL.jpg|thumb|780px|centre| Figure X:<br>'''Inverted microscope composite images of DH5alpha E.coli, serving as negative control'''<br> A: shows YFP fluorescence (514 nm)<br> B: stained with biotinylated anti-YFP antibody and avidin dylight (650 nm)<br> C: Merge of A and B, no signall]] |
| | + | [[File:Team-EPF-Lausanne_POSCTL.jpg|thumb|700px|centre| Figure X:<br>'''Inverted microscope composite images of INP-YFP transformed DH5alpha E.coli'''<br> A: shows YFP fluorescence (514 nm)<br> B: stained with biotinylated anti-YFP antibody and avidin dylight (650 nm)<br> C: Merge of A and B, shows colocalization of fluorescent signal]] |
| | + | |
| | + | |
| | + | '''''Discussion''''' |
| | + | <br> |
| | + | Clearly we see some colocalization of the red and yellow fluorescence with the anti-YFP antibody, but as we were short of time, we weren't able to incubate the antibody long enough, we only incubated for 15 minutes. We will also need to sequence verify the construct to support the colony PCR we did, '''but still results are promising'''. |
| | | | |
| - | ==Results==
| |
| | <html> | | <html> |
| - | In the following section, you will find results concerning our cell surface display work.
| + | <br>Here is the nomenclature we will further use: |
| - | Before moving on it is important to define the nomenclature we will further use:
| + | |
| | <br> | | <br> |
| | 1)Initially used plasmid: | | 1)Initially used plasmid: |
| - | <br>INP_EYFP Biobrick: <a href= "http://parts.igem.org/Part:BBa_K523013">BBa_523013</a> | + | <br>INP_EYFP Biobrick: <a href= "http://parts.igem.org/Part:BBa_K523013">BBa_K523013</a> |
| - | <br>Streptavidin Alive: <a href="http://parts.igem.org/Part:BBa_K1111010">BBa_K1111010</a> | + | <br>Streptavidin Alive: <a href="http://parts.igem.org/Part:BBa_K1111010">BBa_K1111010</a> |
| | <br>Streptavidin Dead: <a href="http://parts.igem.org/Part:BBa_K1111011">BBa_K1111011</a> | | <br>Streptavidin Dead: <a href="http://parts.igem.org/Part:BBa_K1111011">BBa_K1111011</a> |
| - | <br>Streptavidin iGEM: <a href="http://parts.igem.org/Part:BBa_K283010">BBa_K283010</a> | + | <br>Streptavidin iGEM: <a href="http://parts.igem.org/Part:BBa_K283010">BBa_K283010</a>. |
| | | | |
| | <br><br> | | <br><br> |
| | 2) PCR reactions products | | 2) PCR reactions products |
| - | <br>INP_construct = BBa_K523013 after PCR without EYFP; SA = Streptavidin Alive; SD = Streptavidin Dead; SI = Streptavidin iGEM. | + | <br>INP_construct = BBa_K523013 after PCR without EYFP |
| | + | <br>SA = Streptavidin Alive; SD = Streptavidin Dead |
| | + | <br>SI = Streptavidin iGEM. |
| | <br><br> | | <br><br> |
| | | | |
| Line 83: |
Line 296: |
| | <br>ISD = INP_Streptavidin Dead gibson product<a href="http://parts.igem.org/Part:BBa_K1111013">BBa_K1111013</a> | | <br>ISD = INP_Streptavidin Dead gibson product<a href="http://parts.igem.org/Part:BBa_K1111013">BBa_K1111013</a> |
| | <br>ISI = INP_Streptavidin iGEM BBa_K283010 gibson product<a href="http://parts.igem.org/Part:BBa_K1111014">BBa_K1111014</a> | | <br>ISI = INP_Streptavidin iGEM BBa_K283010 gibson product<a href="http://parts.igem.org/Part:BBa_K1111014">BBa_K1111014</a> |
| | + | |
| | </html> | | </html> |
| | + | <br> |
| | + | To learn more about how we would continue, see [https://2013.igem.org/Team:EPF_Lausanne/Next_steps Next Steps] or [https://2013.igem.org/Team:EPF_Lausanne/Perspectives Perspectives] |
| | | | |
| | <br> | | <br> |
| - | <br>Our first experimental steps were to perform Polymerase Chain Reactions (PCR) in order to isolate the our Bricks.
| |
| | | | |
| - | <!--PCR images-->
| + | <br><br> |
| - | [[File:Team-EPF-Lausanne_PCRStrepA.jpeg|thumb|250px|left|Figure x: 0.8% Gel electrophoresis to verify PCR of Streptavidin Alive coding sequence]]
| + | |
| - | [[File:Team-EPF-Lausanne_PCRStrepD.jpeg|thumb|200px|left|Figure y: 0.8% Gel electrophoresis to verify PCR of Streptavidin Dead coding sequence]]
| + | |
| - | [[File:Team-EPF-Lausanne_PCRAtrepI.jpeg|thumb|250px|left|Figure z: 0.8% Gel electrophoresis to verify PCR of Streptavidin iGEM coding sequence]]
| + | |
| - | <br><br><br><br><br><br><br><br><br><br>
| + | |
| | | | |
| - | <!--Gibson Assembly images -->
| + | ==References== |
| - | [[File:Team-EPF-Lausanne_plate_ISA.jpeg|thumb|260px|left|Figure x: 0.8% Gel electrophoresis to verify INP_Streptavidin Alive Gibson Assembly product ]]
| + | |
| - | [[File: Team-EPF-Lausanne_GAINPStrepD.jpg|thumb|210px|left|Figure y: 0.8% Gel electrophoresis to verify INP_Streptavidin Dead Gibson Assembly product]]
| + | |
| - | [[File: Team-EPF-Lausanne_GAINPStrepI.jpg|thumb|260px|left|Figure z: 0.8% Gel electrophoresis to verify INP_Streptavidin iGEM Gibson Assembly product]]
| + | |
| - | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> <br>
| + | |
| | | | |
| - | <br>The previous Gels show that our PCR reaction to amplify coding sequences and the Gibson Assembly reaction to obtain our biobricks worked as expected. | + | [1] Surface display of Zymomonas mobilis levansucrase by using the ice-nucleation protein of Pseudomonas syringae, Jung HC, Lebeault JM, Pan JG. (1998) |
| - | Indeed, we can see characteristic bands corresponding to the right Gibson product. Then another way to verify that our Gibson assembly recation potentially worked was to tranform cells with this product. The transformed cells should be able to grow if they took up the gibson product. The on only problem with this second verification is that the Gibson product could still contain parental plasmid,which insertion in cells can also lead to cells resistance and survival. Here, you are not provided with the transformation plates and their controls because they worked as expected. You can only observe plates containing cells from the glycerol stocks we made of our gibson products .
| + | <br> |
| | + | [2] Identification and purification of a bacterial ice-nucleation protein, Wolber PK, Deininger CA, Southworth MW, Vanderkerckhove J, van Montagu M and Warren GJ (1986) |
| | + | <br> |
| | + | [3] Laboratory of Mark Howarth at Oxford university Department of Biochemistry, for providing us with the cloning plasmids for Streptavidin.<br> |
| | + | [4] Clustering of ice nucleation protein correlates with ice nucleation activity, Mueller GM, Wolber PK, Warren GJ. (1990) |
| | + | <br> |
| | | | |
| - | <!--Plates images-->
| + | </div> |
| - | [[File:Team-EPF-Lausanne_plate_ISA.jpeg|thumb|230px|left|Figure x: INP_Streptavidin Alive Gibson transformed cells plated from the glycerol stock ]]
| + | |
| - | [[File: Team-EPF-Lausanne_plate_ISD.jpeg|thumb|230px|left|Figure y: INP_Streptavidin Dead Gibson transformed cells plated from the glycerol stock]]
| + | |
| - | [[File: Team-EPF-Lausanne_plate_ISI.jpeg|thumb|230px|left|Figure z: INP_Streptavidin iGEM Gibson transformed cells plated from the glycerol stock]]
| + | |
| - | <br><br><br><br><br><br><br><br><br><br>
| + | |
| - | | + | |
| - | We also sent our constructs for sequencing, using VR and VF2 iGEM primers but also another forward primer we designed on ourselves <html><a href="http://parts.igem.org/Part:BBa_K1111012:Experience">results_ISA </a>; <a href="http://parts.igem.org/Part:BBa_K1111013:Experience">results_ISD </a>;<a href="http://parts.igem.org/Part:BBa_K1111014:Experience">results_ISI </a></html>.
| + | |
| - | The sequencing results were also good.
| + | |
| - | From these previous documents, we can simply conclude that our basic cloning strategy was working.
| + | |
| - | Now we needed to test the localisation at the outer membrane and the functionnality of the expressed protein.
| + | |
| - | In order to do so, we designed immunostaining assays to verify localisation at the outer membrane and biotin-staining assay for the protein functionnality (see protocols).
| + | |
| - | Then we visualized the samples using a NIKON Eclipse TI inverted microscope and also a Confocal microscope.
| + | |
| - | | + | |
| - | ==References==
| + | |
| | | | |
| | {{Template:EPFL2013Footer}} | | {{Template:EPFL2013Footer}} |

 "
"