Team:TU Darmstadt/labbook
From 2013.igem.org
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<html> | <html> | ||
| - | |||
<style type="text/css"> | <style type="text/css"> | ||
| Line 16: | Line 15: | ||
{ | { | ||
background-color: white; | background-color: white; | ||
| - | background-image: url("/wiki/images/ | + | background-image: url("/wiki/images/5/5c/Hintergrund_Grün(ohne_Header%2C_Logo_und_Schlagschatten).jpg"); |
background-attachment: scroll; | background-attachment: scroll; | ||
background-position: 50% 0; | background-position: 50% 0; | ||
| Line 27: | Line 26: | ||
{ | { | ||
background: white; | background: white; | ||
| - | background-image: url( | + | background-image: url("/wiki/images/2/2f/Header_%28mit_Logo%29.jpg"); |
margin: 0 auto; | margin: 0 auto; | ||
height:250px ; | height:250px ; | ||
| Line 42: | Line 41: | ||
p {line-height:1.5em; margin:0 0 15px; text-align:justify; color:white;} | p {line-height:1.5em; margin:0 0 15px; text-align:justify; color:white;} | ||
h1 {font-size:1.8em; font-weight:400; margin:0 0 12px;} | h1 {font-size:1.8em; font-weight:400; margin:0 0 12px;} | ||
| + | |||
#mm_icon1 | #mm_icon1 | ||
| Line 108: | Line 108: | ||
{ | { | ||
position:absolute; | position:absolute; | ||
| - | top: | + | top:10px; |
| - | left: | + | left:500px; |
| - | z-index: | + | z-index: 5; |
} | } | ||
| + | |||
ul { | ul { | ||
| Line 208: | Line 209: | ||
<br> | <br> | ||
<br> | <br> | ||
| + | |||
<!-- Taskbar --> | <!-- Taskbar --> | ||
| Line 217: | Line 219: | ||
<img alt="Problem" src="/wiki/images/6/66/Darmstadt_green_Problem.jpg" width="100" height="30"></a> | <img alt="Problem" src="/wiki/images/6/66/Darmstadt_green_Problem.jpg" width="100" height="30"></a> | ||
| - | |||
| - | |||
<a href="https://2013.igem.org/Team:TU_Darmstadt/result"> | <a href="https://2013.igem.org/Team:TU_Darmstadt/result"> | ||
| Line 228: | Line 228: | ||
<a href="https://2013.igem.org/Team:TU_Darmstadt/team"> | <a href="https://2013.igem.org/Team:TU_Darmstadt/team"> | ||
<img alt="team" src="/wiki/images/a/a4/Darmstadt_green_Team.jpg" width="70" height="30"></a> | <img alt="team" src="/wiki/images/a/a4/Darmstadt_green_Team.jpg" width="70" height="30"></a> | ||
| - | |||
<br> | <br> | ||
| Line 241: | Line 240: | ||
<a href="https://2013.igem.org/Team:TU_Darmstadt/labbook"> | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook"> | ||
| - | <img alt="team" src="/wiki/images/ | + | <img alt="team" src="/wiki/images/4/4c/10._Labbook_(angewählt).jpg" width="90" height="30"></a> |
| - | |||
| - | |||
| - | |||
| - | < | + | </div> |
| - | + | ||
<br><br><br><br> | <br><br><br><br> | ||
| + | <center> | ||
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook"> | ||
| + | <font size="8" color="#F0F8FF" face="Arial regular">Lab book |</font> | ||
| + | </a> | ||
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/materials"> | ||
| + | <font size="8" color="#F0F8FF" face="Arial regular">Materials |</font> | ||
| + | </a> | ||
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/protocols"> | ||
| + | <font size="8" color="#F0F8FF" face="Arial regular">Protocols</font> | ||
| + | </a> | ||
| + | </center> | ||
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook/DetectionConstruct | + | |
| + | <br><br> | ||
| + | |||
| + | |||
| + | |||
| + | <h2><font size="6" color="#F0F8FF" face="Arial regular">Detection construct</font></h2> <br> | ||
| + | |||
| + | |||
| + | <font size="3" color="#F0F8FF" face="Arial regular"> | ||
| + | <p text-aligne:left style="margin-left:50px; margin-right:50px"> | ||
| + | |||
| + | The assembly of our detection construct proved to be tricky. Not only the traditional cloning approach failed but also the assembly using synthesized gBlocks delivered no results. We finally turned to an In-Fusion cloning approach to assemble our construct. Visit our lab book to read more about our work flow. | ||
| + | |||
| + | </p></font> | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook/DetectionConstruct"> | ||
<img alt="Labbookdetection" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> | <img alt="Labbookdetection" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> | ||
| - | <font size="6" color="#F0F8FF" face="Arial regular">Visit | + | <font size="6" color="#F0F8FF" face="Arial regular">Visit our lab book for the detection construct</font><br> |
</a> | </a> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | <h2><font size="6" color="#F0F8FF" face="Arial regular"> | + | |
| + | <br><br> | ||
| + | |||
| + | <h2><font size="6" color="#F0F8FF" face="Arial regular">Fluorescence proteins</font></h2> <br> | ||
<font size="3" color="#F0F8FF" face="Arial regular"> | <font size="3" color="#F0F8FF" face="Arial regular"> | ||
<p text-aligne:left style="margin-left:50px; margin-right:50px"> | <p text-aligne:left style="margin-left:50px; margin-right:50px"> | ||
| - | + | The genes for our FRET pair, mKate and LssmOrange, were isolated by PCR, cloned into the biobrick vector and sequenced afterwards. The genes were also cloned into a commercially available expression vector in order to characterize the fluorescent proteins. Visit our lab book to read more about our work flow. | |
| + | <center> | ||
| + | <body> | ||
<br> | <br> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/ | + | |
| - | <img alt=" | + | <img alt="mkate" src="/wiki/images/3/3a/Mkate-active_light1.png" width="50%" height="50%" align="right"> |
| - | <font size="6" color="#F0F8FF" face="Arial regular"> | + | |
| + | <img alt="mkateactive" src="/wiki/images/5/58/Mkate-active_surface_activesite4_light1.png" width="50%" height="50%" align="right"> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | </p></font> | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook/Biobricks"> | ||
| + | <img alt="Labbookbiobricks" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> | ||
| + | <font size="6" color="#F0F8FF" face="Arial regular">Visit our lab book for the fluorescence proteins</font><br> | ||
</a> | </a> | ||
| + | <br> | ||
| + | <h2><font size="6" color="#F0F8FF" face="Arial regular">Safety</font></h2> <br> | ||
| + | |||
| + | |||
| + | <font size="3" color="#F0F8FF" face="Arial regular"> | ||
| + | <p text-aligne:left style="margin-left:50px; margin-right:50px"> | ||
| + | |||
| + | The assembly of the blue-light sensitive pDawn construct from gBlocks failed. Although the construction of the pezT construct from gBlocks was successful, multiple transformations of pSB1C3-pezT failed. Visit our lab book to read more about our work flow. | ||
| + | |||
| + | </p></font> | ||
| + | <br><br> | ||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/safety/Labjournal"> | ||
| + | <img alt="Labbookbiobricks" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> | ||
| + | <font size="6" color="#F0F8FF" face="Arial regular">Visit our lab book for the safety construct</font><br /> | ||
| + | </a> | ||
| | ||
| - | <a href="https://2013.igem.org/Team:TU_Darmstadt/ | + | <h2><font size="6" color="#F0F8FF" face="Arial regular">Cutinase</font></h2> <br> |
| + | |||
| + | |||
| + | <font size="3" color="#F0F8FF" face="Arial regular"> | ||
| + | <p text-aligne:left style="margin-left:50px; margin-right:50px"> | ||
| + | |||
| + | Here, we present the improvement approach of the <i>Fusarium Solani Cutinase</i> (FsC, PID: BBa K808025). | ||
| + | The FsC was one of the PET cleavage enzymes from the iGEM Team TU-Darmstadt 2012. We improved this part | ||
| + | with a pelB leader and a reporter(mRFP1-His6) system with a TEV cleavage site. This construct is regulated by pBAD | ||
| + | (PID: BBa I0500) and cloned into the pSB1C3 backbone. | ||
| + | |||
| + | </p></font> | ||
| + | <center> | ||
| + | <img alt="fsc" src="/wiki/images/0/0d/Fscpet2.png" width="50%" height="50%"> | ||
| + | <br><br> | ||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook/Cutinase"> | ||
<img alt="Labbookdetection" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> | <img alt="Labbookdetection" border="5" width="57,5" height="50" src="/wiki/images/8/81/Book2.gif"> | ||
| - | <font | + | <font size="6" color="#F0F8FF" face="Arial regular">Visit our lab book for the cutinase</font><br /> |
</a> | </a> | ||
| - | |||
Latest revision as of 03:13, 5 October 2013
Detection construct
The assembly of our detection construct proved to be tricky. Not only the traditional cloning approach failed but also the assembly using synthesized gBlocks delivered no results. We finally turned to an In-Fusion cloning approach to assemble our construct. Visit our lab book to read more about our work flow.
 Visit our lab book for the detection construct
Visit our lab book for the detection constructFluorescence proteins
The genes for our FRET pair, mKate and LssmOrange, were isolated by PCR, cloned into the biobrick vector and sequenced afterwards. The genes were also cloned into a commercially available expression vector in order to characterize the fluorescent proteins. Visit our lab book to read more about our work flow.
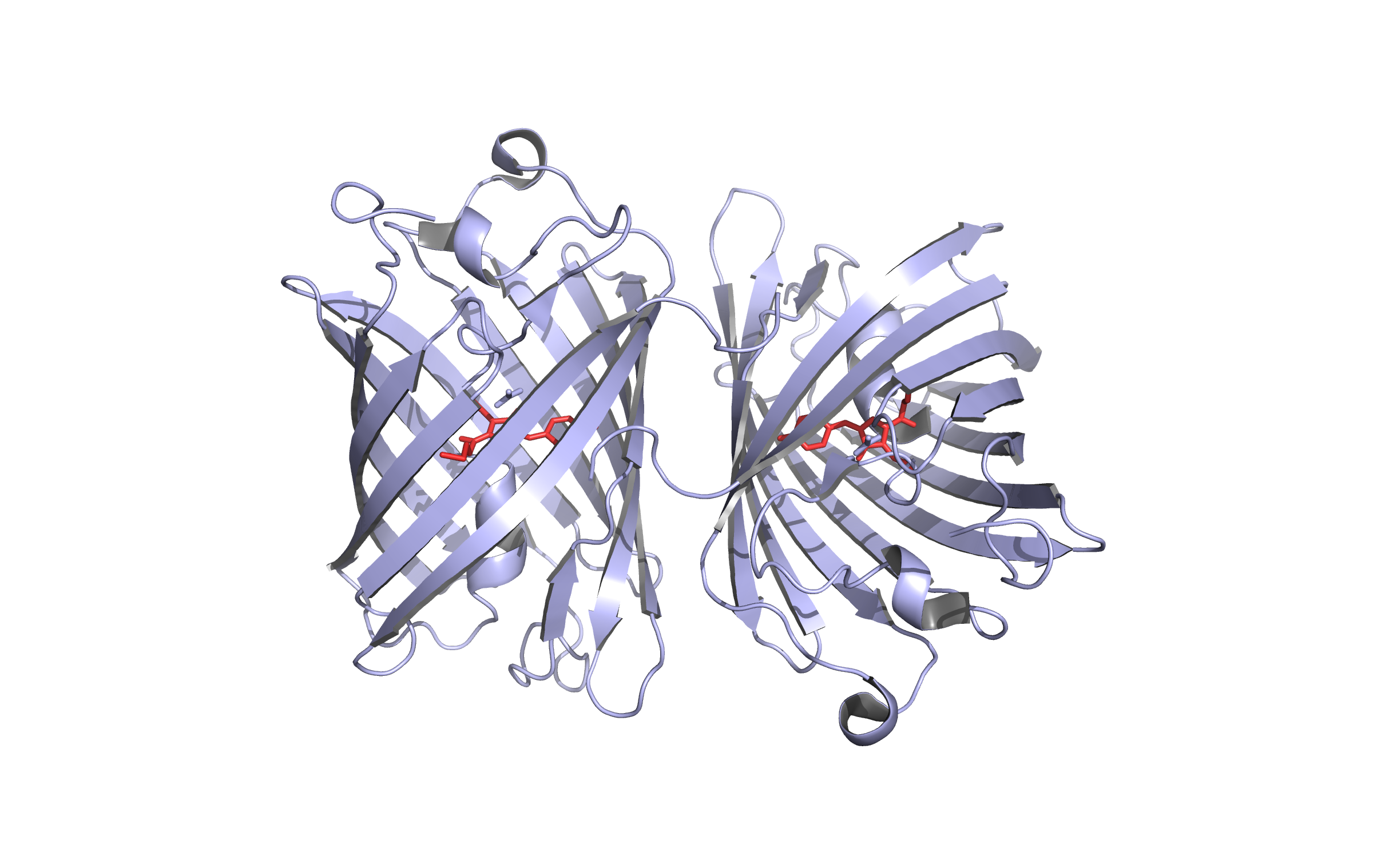
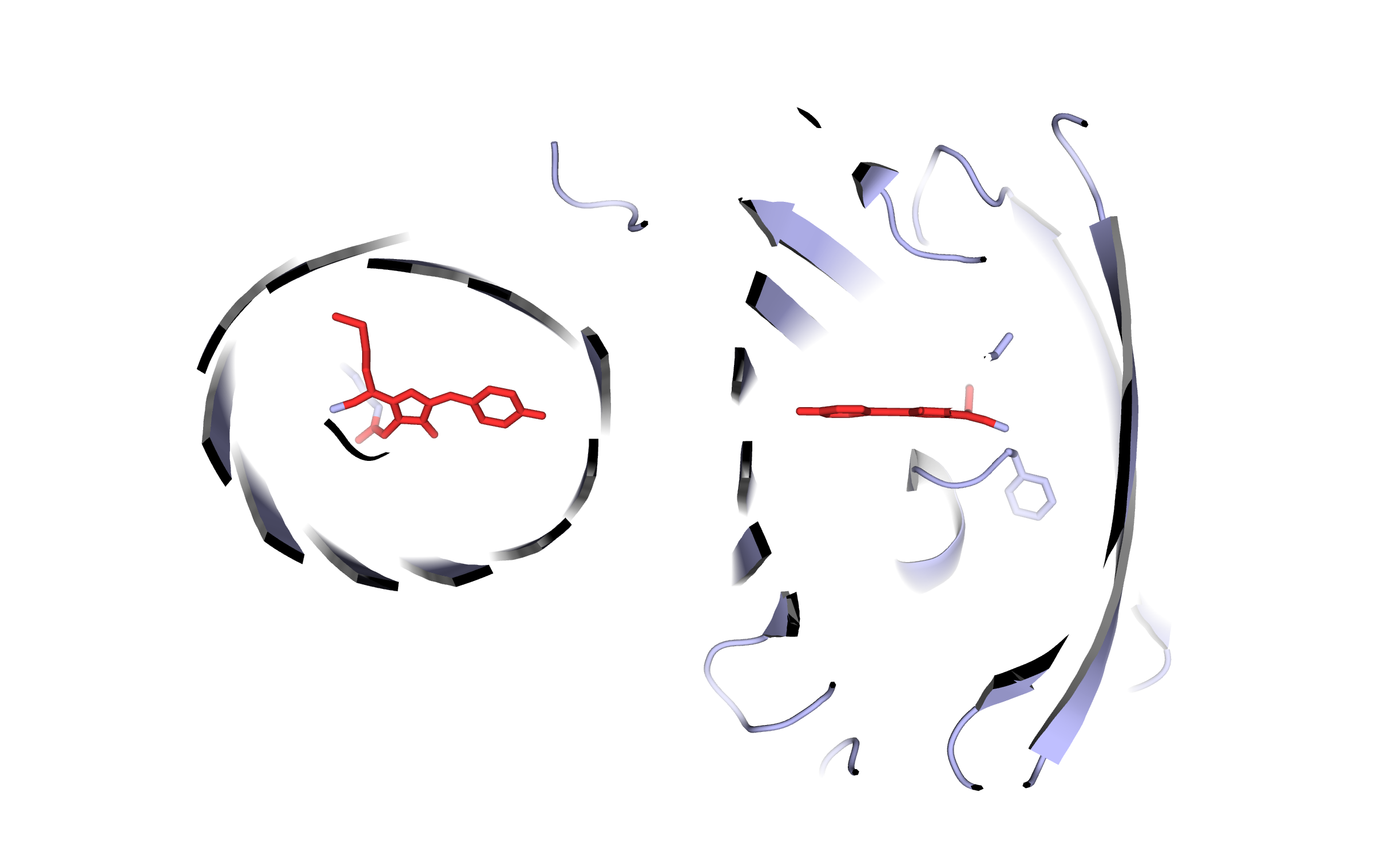
 Visit our lab book for the fluorescence proteins
Visit our lab book for the fluorescence proteinsSafety
The assembly of the blue-light sensitive pDawn construct from gBlocks failed. Although the construction of the pezT construct from gBlocks was successful, multiple transformations of pSB1C3-pezT failed. Visit our lab book to read more about our work flow.
 Visit our lab book for the safety construct
Visit our lab book for the safety constructCutinase
Here, we present the improvement approach of the Fusarium Solani Cutinase (FsC, PID: BBa K808025). The FsC was one of the PET cleavage enzymes from the iGEM Team TU-Darmstadt 2012. We improved this part with a pelB leader and a reporter(mRFP1-His6) system with a TEV cleavage site. This construct is regulated by pBAD (PID: BBa I0500) and cloned into the pSB1C3 backbone.
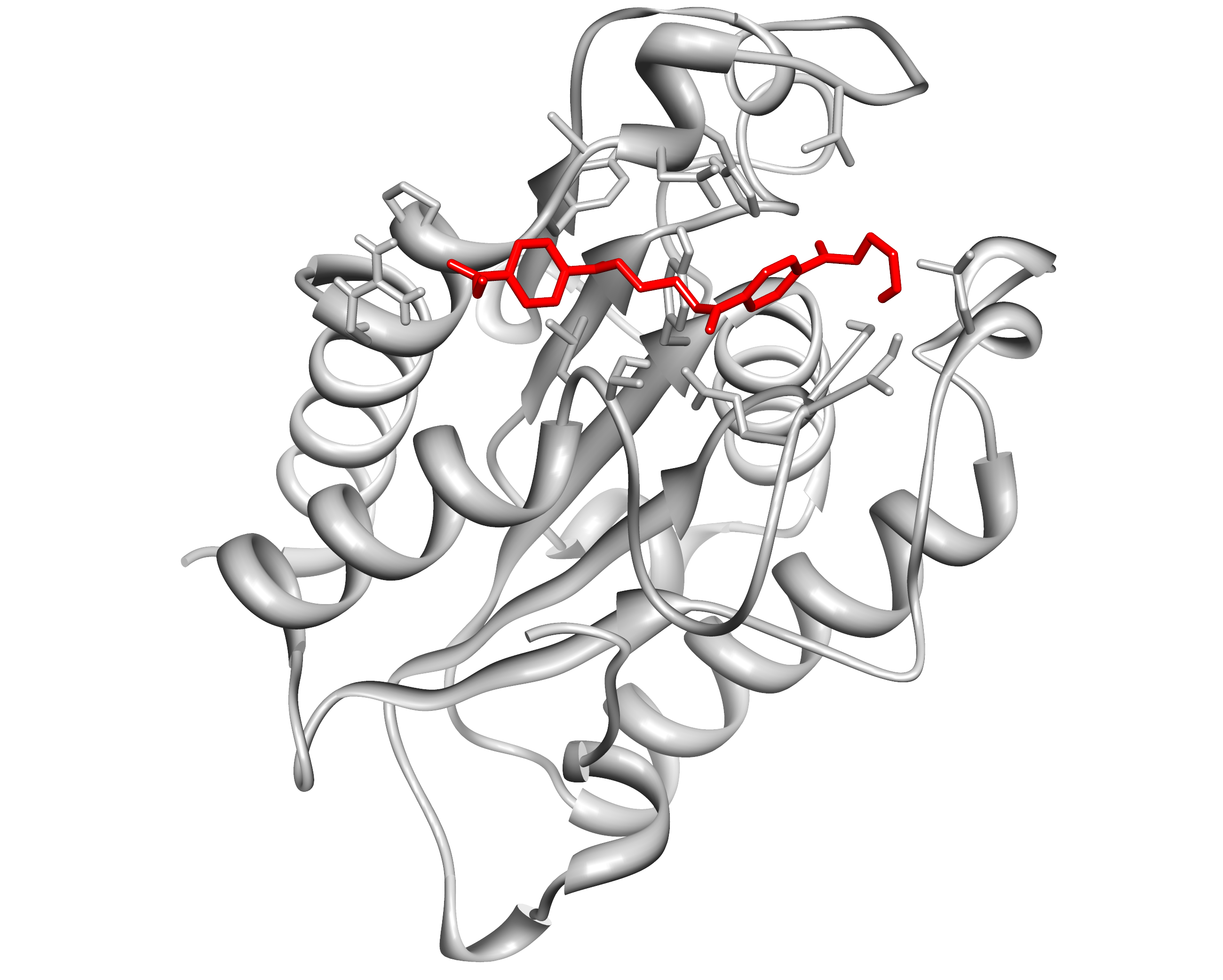
 Visit our lab book for the cutinase
Visit our lab book for the cutinase "
"