Team:Manchester/Parts
From 2013.igem.org
| (10 intermediate revisions not shown) | |||
| Line 80: | Line 80: | ||
position:absolute; | position:absolute; | ||
clear:both; | clear:both; | ||
| - | width: | + | width:960px; |
| - | + | ||
top:120px; | top:120px; | ||
| - | left: | + | left:0; |
| - | + | ||
background-color:white; | background-color:white; | ||
| Line 214: | Line 212: | ||
width:900px; | width:900px; | ||
font-style:Trebuchet MS; | font-style:Trebuchet MS; | ||
| - | font-size: | + | font-size:25px; |
color:#4c0082; | color:#4c0082; | ||
background-color:#F2F2F2; | background-color:#F2F2F2; | ||
| Line 237: | Line 235: | ||
.text3 img | .text3 img | ||
{ | { | ||
| + | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | } | ||
| + | |||
| + | .text4 #footer | ||
| + | { | ||
| + | width:870px; | ||
| + | height:70px; | ||
| + | margin-top:5px; | ||
| + | padding:5px; | ||
| + | |||
-webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
-moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| Line 248: | Line 258: | ||
height:200px; | height:200px; | ||
padding:20px; | padding:20px; | ||
| + | background-color:#f2f2f2; | ||
-webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
-moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| Line 278: | Line 289: | ||
height:300px; | height:300px; | ||
padding:20px; | padding:20px; | ||
| + | font-style:Trebuchet MS; | ||
| + | font-size:14px; | ||
| + | color:#4c0082; | ||
| + | background-color:#f2f2f2; | ||
-webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
-moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| Line 341: | Line 356: | ||
.text4 | .text4 | ||
{ | { | ||
| + | background-color:#F2F2F2; | ||
width:880px; | width:880px; | ||
margin:5px 20px; | margin:5px 20px; | ||
| - | height: | + | height:1300px; |
padding:20px; | padding:20px; | ||
-webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| Line 354: | Line 370: | ||
float:left; | float:left; | ||
margin-right:20px; | margin-right:20px; | ||
| + | background-color:#F2F2F2; | ||
-webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
-moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| Line 362: | Line 379: | ||
{ | { | ||
float:left; | float:left; | ||
| - | + | background-color:#F2F2F2; | |
-webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
-moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| Line 371: | Line 388: | ||
{ | { | ||
float:left; | float:left; | ||
| - | width: | + | width:860px; |
padding:10px; | padding:10px; | ||
font-style:Trebuchet MS; | font-style:Trebuchet MS; | ||
| Line 377: | Line 394: | ||
color:#4c0082; | color:#4c0082; | ||
background-color:#F2F2F2; | background-color:#F2F2F2; | ||
| + | |||
| + | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | } | ||
| + | |||
| + | .text6 | ||
| + | { | ||
| + | background-color:#F2F2F2; | ||
| + | width:880px; | ||
| + | margin:5px 20px; | ||
| + | height:730px; | ||
| + | padding:20px; | ||
| + | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | } | ||
| + | |||
| + | .text6 #left | ||
| + | { | ||
| + | float:left; | ||
| + | margin-right:20px; | ||
| + | background-color:#F2F2F2; | ||
| + | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | } | ||
| + | |||
| + | .text6 #right | ||
| + | { | ||
| + | float:left; | ||
| + | background-color:#F2F2F2; | ||
| + | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | } | ||
| + | |||
| + | .text6 p | ||
| + | { | ||
| + | float:left; | ||
| + | width:860px; | ||
| + | padding:10px; | ||
| + | font-style:Trebuchet MS; | ||
| + | font-size:14px; | ||
| + | color:#4c0082; | ||
| + | background-color:#F2F2F2; | ||
| + | |||
| + | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | -moz-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| + | } | ||
| + | |||
| + | .text6 #footer | ||
| + | { | ||
| + | width:870px; | ||
| + | height:30px; | ||
| + | margin-top:5px; | ||
| + | padding:5px; | ||
-webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | -webkit-box-shadow: 0px 0px 5px 0px rgba(0,0,0,0.75); | ||
| Line 530: | Line 605: | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
<div class="text3"> | <div class="text3"> | ||
| + | <center><i>We used an existing BioBrick component with Constitutive promoter and RBS<br><br> (BBa_K608002) to express our BioBrick parts:</i></center><br> | ||
<img src="https://static.igem.org/mediawiki/2013/1/1a/Expression_vectors.png" width="900" height="300"/> | <img src="https://static.igem.org/mediawiki/2013/1/1a/Expression_vectors.png" width="900" height="300"/> | ||
<!--<p id="footer"><b>Figure 5. Overlay of structures from 1 ns FabA simulation. Images of overlaid from the following respective time points: 0 ps, 250 ps, 500 ps, 750 ps and 1000 ps with the following colours indicating each individual image: Green, Blue, Purple, Orange and Grey, respectively. Both the N-Terminal and C-Terminal, are specified (Dotted Box), with a zoom in on each respective terminal at an angle appropriate to visualise the positions of the terminals. | <!--<p id="footer"><b>Figure 5. Overlay of structures from 1 ns FabA simulation. Images of overlaid from the following respective time points: 0 ps, 250 ps, 500 ps, 750 ps and 1000 ps with the following colours indicating each individual image: Green, Blue, Purple, Orange and Grey, respectively. Both the N-Terminal and C-Terminal, are specified (Dotted Box), with a zoom in on each respective terminal at an angle appropriate to visualise the positions of the terminals. | ||
| Line 545: | Line 617: | ||
<!--<p id="footer"><b>Figure 1. Two colour image of FabA homodimer with labelled N- and C-Termini (arrows)</b><br>--> | <!--<p id="footer"><b>Figure 1. Two colour image of FabA homodimer with labelled N- and C-Termini (arrows)</b><br>--> | ||
<p><b>Successful Expression of delta 9 and delta 12 desaturase, and FabA</b><br> | <p><b>Successful Expression of delta 9 and delta 12 desaturase, and FabA</b><br> | ||
| - | <p>In order to characterise our biobricks, we made use of the standard parts found in the registry. By inserting the created biobricks BBa_K1027001 and BBa_K1027002 in to BBa_K608002 (a biobrick consisting of a ribosomal binding site and a constitutive promoter), we were able to create a new construct that expressed the delta 9 desaturase | + | <p>In order to characterise our biobricks, we made use of the standard parts found in the registry. By inserting the created biobricks BBa_K1027001 and BBa_K1027002 in to BBa_K608002 (a biobrick consisting of a ribosomal binding site and a constitutive promoter), we were able to create a new construct that expressed the delta 9 desaturase, delta 12 desaturase and FabA proteins. The constructs are shown above.</p> |
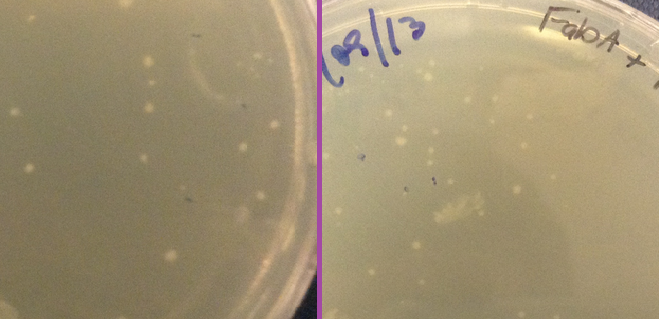
<p><b>Plates</b></p> | <p><b>Plates</b></p> | ||
| - | <p>Another piece of evidence suggesting that our constructs were successfully expressing our delta 9 desaturase and delta 12 desaturase enzymes was the size of the colonies grown on the plates. Pictures of the plates are shown below. On the far left is a control plate, with bacteria transformed with BBa_K608002 but with no gene inserted in front of the promoter. To the right of that are our delta 12 colonies. They are much smaller than the control, and even taking 20 hours to grow to that size as opposed to the usual 16 hours. We hypothesise that because delta 12 desaturase is a membrane-bound protein that the <i>E. coli</i> does not normally express, constitutively expressing it could inhibit the bacteria and slow growth. Compare this to | + | <p>Another piece of evidence suggesting that our constructs were successfully expressing our delta 9 desaturase and delta 12 desaturase enzymes was the size of the colonies grown on the plates. Pictures of the plates are shown below. On the far left is a control plate, with bacteria transformed with BBa_K608002 but with no gene inserted in front of the promoter. To the right of that are our delta 12 colonies. They are much smaller than the control, and even taking 20 hours to grow to that size as opposed to the usual 16 hours. We hypothesise that because delta 12 desaturase is a membrane-bound protein that the <i>E. coli</i> does not normally express, constitutively expressing it could inhibit the bacteria and slow growth. Compare this to FabA on the far right. Looking through the literature, we found that overexpression of FabA results in no significant difference in growth size or speed relative to the wild type strain (Luo et al, 2009). Compared to the control plate, we also found that the colonies grew normally.</p> |
<p><b>Gel Digests</b></p> | <p><b>Gel Digests</b></p> | ||
| Line 554: | Line 626: | ||
<b>delta 9 desaturase:</b> Cut with BamHI + EcoRV. Expected bands 1424 bp, 1262 bp, 99 bp.<br> | <b>delta 9 desaturase:</b> Cut with BamHI + EcoRV. Expected bands 1424 bp, 1262 bp, 99 bp.<br> | ||
<b>delta 12 desaturase:</b> Cut with BamHI, XbaI, PstI. Expected bands 2127 bp, 656 bp, 430 bp.<br> | <b>delta 12 desaturase:</b> Cut with BamHI, XbaI, PstI. Expected bands 2127 bp, 656 bp, 430 bp.<br> | ||
| - | <b>FabA</b> Cut with EcoRV. Expected bands 1429 bp, 1115 bp. Cut with EcoRV, PstI. Expected bands 1153 bp, 1115 bp, 339 bp.</p> | + | <b>FabA</b> Cut with EcoRV. Expected bands 1429 bp, 1115 bp. Cut with EcoRV, PstI. Expected bands 1153 bp, 1115 bp, 339 bp.<br></p> |
<br> | <br> | ||
<p><b>Improvement of existing BioBrick</b></p> | <p><b>Improvement of existing BioBrick</b></p> | ||
<p>Our biobrick BBa_K1027002 (<b>delta 12 desaturase</b>) is an improvement of the <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K925000" target="_blank">BBa_K925000</a> biobrick created by the St Andrews team in 2012. We improved this biobrick from the ground up, starting by taking the protein sequence from <i>Synechocystis</i> sp. PCC 6803. This protein sequence was reverse translated and then codon optimised for expression in <i>E. coli</i>. We believe our redesigned biobrick has two features that the original biobrick does not. Firstly, the original biobrick could not be transformed by iGEM HQ, whereas we have managed successful transformation (as shown by the plate pictures). Next, the St. Andrews biobrick was cloned from cyanobacteria, and so is not optimised for expression in <i>E. coli</i>. As St. Andrews then went on to express their delta 12 desaturase biobrick in <i>E. coli</i> it follows that a sequence optimised for expression in this chassis, as ours was, would perform better than the sequence naturally found in cyanobacteria.</p> | <p>Our biobrick BBa_K1027002 (<b>delta 12 desaturase</b>) is an improvement of the <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K925000" target="_blank">BBa_K925000</a> biobrick created by the St Andrews team in 2012. We improved this biobrick from the ground up, starting by taking the protein sequence from <i>Synechocystis</i> sp. PCC 6803. This protein sequence was reverse translated and then codon optimised for expression in <i>E. coli</i>. We believe our redesigned biobrick has two features that the original biobrick does not. Firstly, the original biobrick could not be transformed by iGEM HQ, whereas we have managed successful transformation (as shown by the plate pictures). Next, the St. Andrews biobrick was cloned from cyanobacteria, and so is not optimised for expression in <i>E. coli</i>. As St. Andrews then went on to express their delta 12 desaturase biobrick in <i>E. coli</i> it follows that a sequence optimised for expression in this chassis, as ours was, would perform better than the sequence naturally found in cyanobacteria.</p> | ||
| - | |||
</p> | </p> | ||
</div> | </div> | ||
| - | + | ||
| - | <img id="left" src="https://static.igem.org/mediawiki/2013/ | + | <div class="double2"> |
| - | <img id="right" src="https://static.igem.org/mediawiki/2013/ | + | <div class="left"> |
| + | <img id="left" src="https://static.igem.org/mediawiki/2013/6/66/Delta12andwtzoomed.png" width="400" height="200"/> | ||
| + | <p id="footer"><b>LB-Agar (CmR) plates after 20h growth at 37°C. Left: Control plate (transformed with empty vector BBa_K608002). Right: Transformed with delta 12 desaturase (note smaller colonies)</b><br> | ||
| + | </div> | ||
| + | |||
| + | <div class="right"> | ||
| + | <img id="right" src="https://static.igem.org/mediawiki/2013/2/28/Fabaandwtzoomed.png" width="400" height="200"/> | ||
| + | <p id="footer"><b>LB-Agar (CmR) plates after 20h growth at 37°C. Left: Control plate (transformed with empty vector BBa_K608002). Right: Transformed with FabA gene (note similar sized colonies)</b><br> | ||
| + | </div> | ||
| + | |||
</div> | </div> | ||
| Line 591: | Line 671: | ||
<img id="left" src="https://static.igem.org/mediawiki/2013/4/40/Orbitrapchromatogramsmanchester.png" width="430" height="450"/> | <img id="left" src="https://static.igem.org/mediawiki/2013/4/40/Orbitrapchromatogramsmanchester.png" width="430" height="450"/> | ||
| - | <img id="right" src="https://static.igem.org/mediawiki/2013/f/f7/Orbitraplcmsflow.png" width="430" height=450"/> | + | <img id="right" src="https://static.igem.org/mediawiki/2013/f/f7/Orbitraplcmsflow.png" width="430" height="450"/> |
| - | + | <p id="footer"><b>Above Left: Chromatograms obtained through Orbitrap LC-MS analysis.<br> Above Right: Workflow of how we went from samples to results<br>Below Left: Bar chart showing increase in linoleic acid when delta 12 desaturase is present<br>Below Right: Heat map showing metabolite abundances </b><br> | |
| - | < | + | <br><br> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | <br> | + | |
| - | <br> | + | |
| - | <br> | + | |
<div class="text5"> | <div class="text5"> | ||
| Line 605: | Line 679: | ||
<img id="right" src="https://static.igem.org/mediawiki/2013/0/0d/Orbitrapheatmapmanchester.png" width="430" height=450"/> | <img id="right" src="https://static.igem.org/mediawiki/2013/0/0d/Orbitrapheatmapmanchester.png" width="430" height=450"/> | ||
| + | |||
| + | |||
| + | |||
| + | <p><b>Further analysis</b><br> | ||
| + | The data obtained from the LC-MS was enormous. 2721 metabolites were detected in the samples, and so we filtered this down to the 43 fatty acids and phospholipids incorporating the compounds we were interested in (oleic acid and linoleic acid). A heat map was generated (seen above) showing the abundances of these 43 phospholipids and fatty acids in the 17 samples we run on the LC-MS. From this heatmap the diversity of detected compounds and high dynamic range can be seen. For the characterisation of our delta 12 desaturase BioBrick construct, a focus on the individual compounds of high intensity was necessary. An example of this can be seen in the bar chart above. Here you can see that, when fed with oleic acid (to a total w/v concentration of 0.1% and 0.5%), the amount of linoleic acid found within this representative phospholipid is much higher in the delta 12 desaturase expression strain than in the wild-type DH5-alpha strain. From this data we can confidently conclude that the delta 12 desaturase is converting the substrate oleic acid into the linoleic acid. Currently, the linoleic is incorporated into a phospholipid, but expression of the tesA gene (biobricked previously) would cleave the compound away to give free fatty acid. <br> | ||
| + | <br> | ||
| + | <b>A work flow of the methods we used to get from sample to data analysis can be seen above.</b> | ||
| + | </p> | ||
</div> | </div> | ||
| - | </div> | + | </div> |
| - | + | ||
| - | </ | + | <div class="text6"> |
| - | + | <img id="left" src="https://static.igem.org/mediawiki/2013/5/53/Wtafmmanc.jpg" width="430" height="450"/> | |
| + | |||
| + | <img id="right" src="https://static.igem.org/mediawiki/2013/9/95/Delta12afmmanc.jpg" width="430" height="450"/> | ||
| + | <p id="footer"><b>Atomic Force Microscopy pictures. Left: WT DH5-alpha. Right: DH5-alpha transformed with delta 12 desaturase expression plasmid.</b><br> | ||
| + | <br><br> | ||
| + | |||
| + | |||
| + | <p><b>Atomic Force Microscopy</b><br> | ||
| + | Whilst overexpression of the delta 12 desaturase led to a 4 fold increase in linoleic acid production incorporated in certain phospholipids, we had hypothesised that both overexpression of a non-native membrane protein and the resulting fatty acid composition changes would possibly alter <i>E. coli</i> membrane structure and potentially interfere growth and replication. To address this, we performed Atomic Force Microscopy (AFM) on transformed wild-type and <i>E. coli</i> transformed with our delta 12 desaturase expression construct. From this we found that there was no significant difference in the membranes between the wild-type and delta 12 desaturase expression constructs. From this we can conclude that overexpression of the non-native membrane bound delta 12 desaturase using our constitutive promoter system does not significantly alter the membranes. Therefore on an industrial scale the expression construct could be modified, such as by the addition of a stronger promoter sequence and RBS site or potentially using inducible expression systems, thus resulting in a greater production of linoleic acid. | ||
| + | </p> | ||
| + | |||
| + | <div class="leftbar"> | ||
<div class="block1"> | <div class="block1"> | ||
<a href="https://2013.igem.org/Team:Manchester/Project">PROJECT</a> | <a href="https://2013.igem.org/Team:Manchester/Project">PROJECT</a> | ||
| Line 645: | Line 738: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
</div> | </div> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 15:53, 28 October 2013
(BBa_K608002) to express our BioBrick parts:


Successful Expression of delta 9 and delta 12 desaturase, and FabA
In order to characterise our biobricks, we made use of the standard parts found in the registry. By inserting the created biobricks BBa_K1027001 and BBa_K1027002 in to BBa_K608002 (a biobrick consisting of a ribosomal binding site and a constitutive promoter), we were able to create a new construct that expressed the delta 9 desaturase, delta 12 desaturase and FabA proteins. The constructs are shown above.
Plates
Another piece of evidence suggesting that our constructs were successfully expressing our delta 9 desaturase and delta 12 desaturase enzymes was the size of the colonies grown on the plates. Pictures of the plates are shown below. On the far left is a control plate, with bacteria transformed with BBa_K608002 but with no gene inserted in front of the promoter. To the right of that are our delta 12 colonies. They are much smaller than the control, and even taking 20 hours to grow to that size as opposed to the usual 16 hours. We hypothesise that because delta 12 desaturase is a membrane-bound protein that the E. coli does not normally express, constitutively expressing it could inhibit the bacteria and slow growth. Compare this to FabA on the far right. Looking through the literature, we found that overexpression of FabA results in no significant difference in growth size or speed relative to the wild type strain (Luo et al, 2009). Compared to the control plate, we also found that the colonies grew normally.
Gel Digests
To confirm that our desired genes were in fact within the expression construct mentioned above, we carried out test digests of our ligated plasmids. Happily, when we ran our digests on an agarose gel, we saw all of the bands we would predict from the expected fragments. The gel pictures are on the right.
delta 9 desaturase: Cut with BamHI + EcoRV. Expected bands 1424 bp, 1262 bp, 99 bp.
delta 12 desaturase: Cut with BamHI, XbaI, PstI. Expected bands 2127 bp, 656 bp, 430 bp.
FabA Cut with EcoRV. Expected bands 1429 bp, 1115 bp. Cut with EcoRV, PstI. Expected bands 1153 bp, 1115 bp, 339 bp.
Improvement of existing BioBrick
Our biobrick BBa_K1027002 (delta 12 desaturase) is an improvement of the BBa_K925000 biobrick created by the St Andrews team in 2012. We improved this biobrick from the ground up, starting by taking the protein sequence from Synechocystis sp. PCC 6803. This protein sequence was reverse translated and then codon optimised for expression in E. coli. We believe our redesigned biobrick has two features that the original biobrick does not. Firstly, the original biobrick could not be transformed by iGEM HQ, whereas we have managed successful transformation (as shown by the plate pictures). Next, the St. Andrews biobrick was cloned from cyanobacteria, and so is not optimised for expression in E. coli. As St. Andrews then went on to express their delta 12 desaturase biobrick in E. coli it follows that a sequence optimised for expression in this chassis, as ours was, would perform better than the sequence naturally found in cyanobacteria.

Orbitrap LC-MS analysis
Delta 9 desaturase and delta 12 desaturase enzymes were chosen because their products, when expressed in their host organism (Synechocystis sp. PCC 6803), convert stearic acid into oleic acid, and oleic acid into linoleic acid respectively. Therefore, we fed batches of transformed DH5-alpha with 2 different concentrations of exogenous fatty acid (0.1% and 0.5% stearic acid fed to the delta 9 desaturase batch, and 0.1% and 0.5% oleic acid fed to the delta 12 desaturase batch), left the cultures growing overnight and then harvested the cells.
To analyse the metabolites extracted from both wild-type DH5-alpha and DH5-alpha expressing our delta 9 desaturase and delta 12 desaturase enzymes, we made use of the MIB’s in-house Orbitrap Liquid Chromatography - Mass Spectrometry (LC-MS). This technique was chosen because of its high mass accuracy and sensitivity. Upon analysing the most abundant metabolites extracted from our expression strains and comparing this data with the most abundant metabolites extracted from wild-type, it is apparent that a massive increase in linoleic acid (incorporated in phosphatidylethanolamines, PE) has occurred. This is demonstrated in the two figures directly below. The chromatograms produced for the delta 12 desaturase expression strains are also shown below. There is a clear difference between the wild-type E. coli fed with exogenous substrate compared with the E. coli strains expressing delta 12 desaturase. It is probable that the peak appearing around 7.9 min in the delta 12 desaturase strains is due to phospholipid incorporating 18:2 (9Z, 12Z) - linoleic acid.






Further analysis
The data obtained from the LC-MS was enormous. 2721 metabolites were detected in the samples, and so we filtered this down to the 43 fatty acids and phospholipids incorporating the compounds we were interested in (oleic acid and linoleic acid). A heat map was generated (seen above) showing the abundances of these 43 phospholipids and fatty acids in the 17 samples we run on the LC-MS. From this heatmap the diversity of detected compounds and high dynamic range can be seen. For the characterisation of our delta 12 desaturase BioBrick construct, a focus on the individual compounds of high intensity was necessary. An example of this can be seen in the bar chart above. Here you can see that, when fed with oleic acid (to a total w/v concentration of 0.1% and 0.5%), the amount of linoleic acid found within this representative phospholipid is much higher in the delta 12 desaturase expression strain than in the wild-type DH5-alpha strain. From this data we can confidently conclude that the delta 12 desaturase is converting the substrate oleic acid into the linoleic acid. Currently, the linoleic is incorporated into a phospholipid, but expression of the tesA gene (biobricked previously) would cleave the compound away to give free fatty acid.
A work flow of the methods we used to get from sample to data analysis can be seen above.


Atomic Force Microscopy
Whilst overexpression of the delta 12 desaturase led to a 4 fold increase in linoleic acid production incorporated in certain phospholipids, we had hypothesised that both overexpression of a non-native membrane protein and the resulting fatty acid composition changes would possibly alter E. coli membrane structure and potentially interfere growth and replication. To address this, we performed Atomic Force Microscopy (AFM) on transformed wild-type and E. coli transformed with our delta 12 desaturase expression construct. From this we found that there was no significant difference in the membranes between the wild-type and delta 12 desaturase expression constructs. From this we can conclude that overexpression of the non-native membrane bound delta 12 desaturase using our constitutive promoter system does not significantly alter the membranes. Therefore on an industrial scale the expression construct could be modified, such as by the addition of a stronger promoter sequence and RBS site or potentially using inducible expression systems, thus resulting in a greater production of linoleic acid.
 "
"







