Team:Tianjin/Project/Alk-Selector&DirectEvolution
From 2013.igem.org
(Created page with "<html> </div> <style type="text/css"> *{margin:0;padding:0;font-family:"微软雅黑","Arial";} body{ width: 960px; height:auto; margin: 0 auto; font-size:12px; font-family:A...") |
Zengbouxan (Talk | contribs) (→Result) |
||
| (38 intermediate revisions not shown) | |||
| Line 226: | Line 226: | ||
<ul> | <ul> | ||
<li> | <li> | ||
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/Introduction # | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Introduction #Background">Background</a> |
</li> | </li> | ||
<li> | <li> | ||
| Line 235: | Line 235: | ||
</li> | </li> | ||
<li> | <li> | ||
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/Introduction #Supplement_information"> | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Introduction #Supplement_information">Supplement Information</a> |
</li> | </li> | ||
</ul> | </ul> | ||
| Line 263: | Line 263: | ||
</li> | </li> | ||
| - | </ul> | + | </ul> |
</li> | </li> | ||
<li class="hmain"> | <li class="hmain"> | ||
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/Alk-Selector&DirectEvolution">Alk-Selector | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Alk-Selector&DirectEvolution">Alk-Selector & Direct Evolution</a> |
| - | + | <ul> | |
<li> | <li> | ||
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/Alk-Selector&DirectEvolution # | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Alk-Selector&DirectEvolution #Design_and_Construction_of_Alk-Selector">Design and Construction of Alk-Selector</a> |
</li> | </li> | ||
<li> | <li> | ||
| Line 289: | Line 289: | ||
<ul> | <ul> | ||
<li> | <li> | ||
| - | <a href="#"> | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Futurework #Summary">Summary</a> |
</li> | </li> | ||
<li> | <li> | ||
| - | <a href="#"> | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Futurework #Future_work">Future work</a> |
</li> | </li> | ||
</ul> | </ul> | ||
| Line 310: | Line 310: | ||
<br/> | <br/> | ||
| - | <center><span style="font-family:Arial;font-size:42px;color:#000;"> Alk-Selector | + | <center><span style="font-family:Arial;font-size:42px;color:#000;"> Alk-Selector and Direct Evolution </span></center> |
<br/> | <br/> | ||
<div style="background-color:#fafafa;width:710px;padding:5px 5px 5px 5px;margin-left:10px;"> | <div style="background-color:#fafafa;width:710px;padding:5px 5px 5px 5px;margin-left:10px;"> | ||
| - | <p> After characterization of Alk-Sensor, we have proved that the AlkR-PalkM regulation system can recognize alkane produced by engineered strain and work well. With this property, we can select the | + | <p> After characterization of Alk-Sensor, we have proved that the AlkR-PalkM regulation system can recognize alkane produced by engineered strain and work well. With this property, we can select out the strains with high productivity of alkane easily according to the output reporter of AlkR-PalkM system. So in this session, we want to build a module called Alk-Selector to help us optimize the alkane biosynthetic pathway of engineered <i>E. coli</i>. </p> |
| - | <p>In our project, we choose NPDC gene, the key gene for alkane biosynthesis, as our target. We will randomly introduce mutation to the gene by error-prone PCR, and build a big library of the NPDC. Then, we will use | + | <p>In our project, we choose NPDC gene, the key gene for alkane biosynthesis, as our target. We will randomly introduce mutation to the gene by error-prone PCR, and build a big library of the NPDC. Then, we will use our Alk-Selector to help us select out the best gene which can give <i>E. coli</i> a high productivity of alkane. </p> |
</div> | </div> | ||
| Line 325: | Line 325: | ||
<br> | <br> | ||
| - | Firstly, we will design a new device Alk-Selector. Although Alk-Sensor can also be regarded as a selector, we don’t want to select just | + | Firstly, we will design a new device Alk-Selector. Although Alk-Sensor can also be regarded as a selector, we don’t want to select just by our eyes which is a tough work. Instead, the special function of antibiotic resistant gene will help us select out the gene with better property automatically in the condition of antibiotic pressure. |
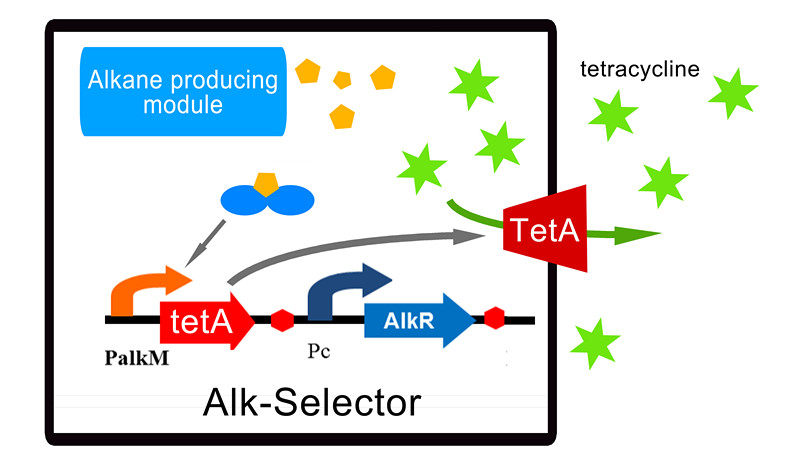
| - | So based on the construction of Alk-Sensor, we | + | So based on the construction of Alk-Sensor, we replaced the RFP reporter with tetracycline resistant gene TetA which is a transporter to pump tetracycline outward the cell. Other components of the Alk-Selector are same to Alk-Sensor. The final construction of Alk-Selector and its working mechanism can be seen in Figure 1. The alkane biosynthesis module expresses NPDC and AAR to produce alkane in E. coli, and then the alkane can activate the expression of tetA through AlkR-PalkM system, and the transporter TetA will give tetracycline resistance to E. coli and make it survive in the environment with tetracycline. |
| + | |||
| + | [[Image:TJU-PROJECT-A1.jpg|thumb|500px|center|<b>Figure 1.</b> Working mechanism of Alk-Selector. Cells cultured in 3ml M9 medium with 12ug/ml tetracycline. When alkanes induce the Alk-Selector, protein tetA will express, and make cells survive in the environment of tetracycline.]] | ||
| - | |||
<br> | <br> | ||
| Line 335: | Line 336: | ||
<br> | <br> | ||
| - | To verify | + | To verify that Alk-Selector will generate more tetracycline resistance when more alkanes are produced, we cotransformed the Alk-Selector and an alkane producing module to the <i>E. coli</i> BL21. The control strain just had Alk-Selector. These two strains were cultured in M9 medium with 12ug/ml tetracycline. After 48h, we can find the growth of strain with alkane producing module was higher than the control just as Figure 2 showed. |
| - | It | + | It proved that our Alk-Selector can work. |
| + | |||
| + | [[Image:TJU-project-D4-2.jpg|thumb|500px|center|<b>Figure 2.</b> Construction scheme of two Alk-Selector strains.Strain one contains alkane producer part(left),the other contains none-producer part (right) .And the result is upon the structure. The none-producer strain grew much slower than alkane producing one.]] | ||
| - | |||
<br> | <br> | ||
| Line 345: | Line 347: | ||
<br> | <br> | ||
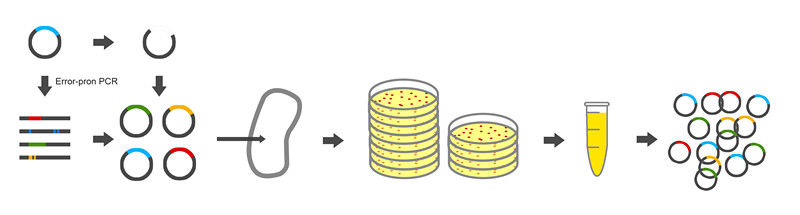
| - | For library construction, we reconstructed the alkane producing | + | For library construction, we reconstructed the alkane producing module of low productivity by adding restriction site BamHI and SacI at the two ends of gene NPDC for library construction. The workflow of the library construction is showed in Figure 3. |
| - | [[Image: | + | [[Image:TJU-project-A3.jpg|thumb|600px|center|<b>Figure 3.</b> The workflow of the library construction.]] |
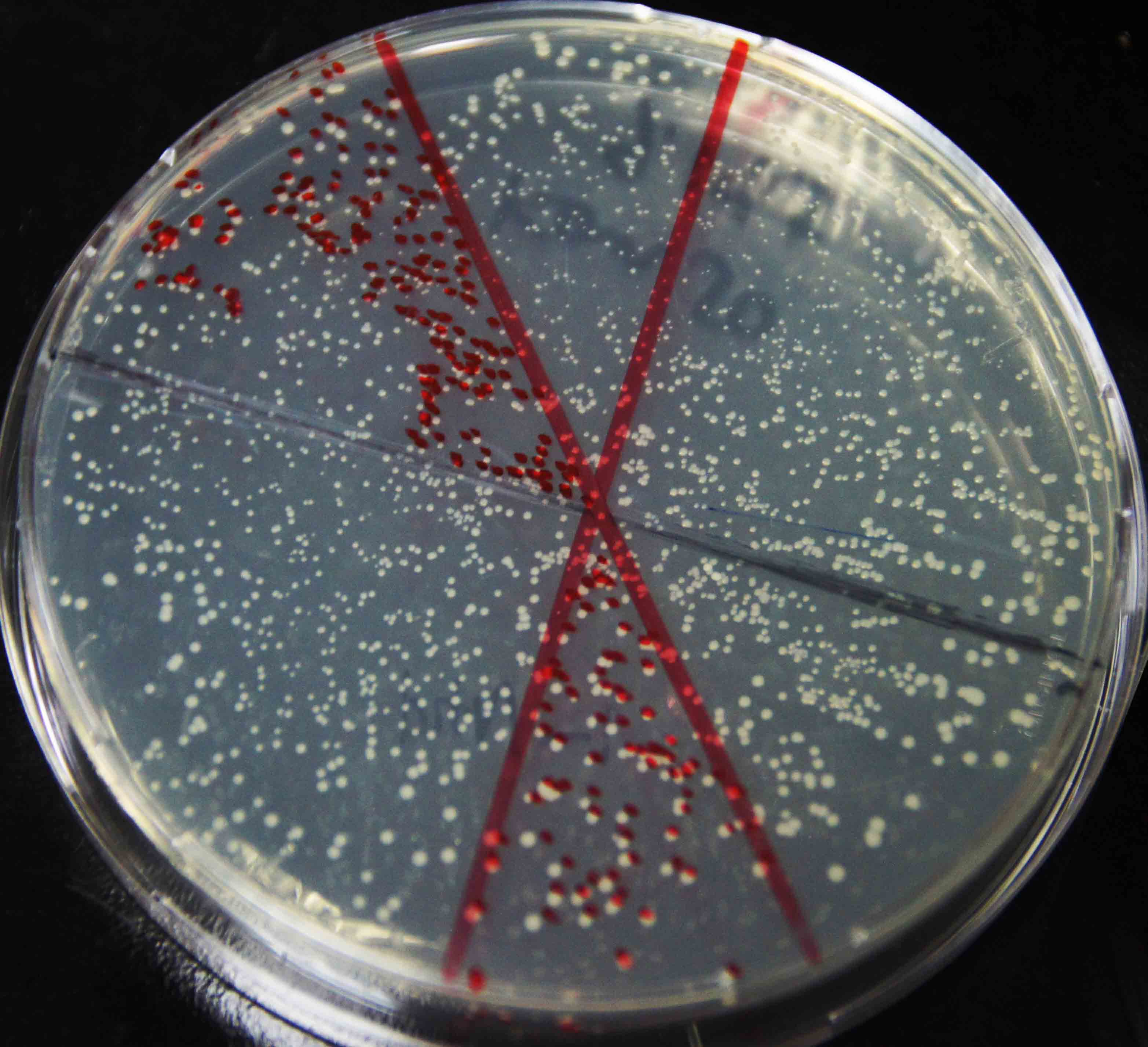
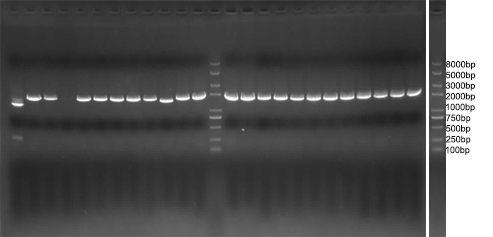
| - | We amplified NPDC by error-prone PCR, and ligand on the plasmid. The | + | We amplified NPDC by error-prone PCR, and ligand on the plasmid. The product of ligation is transformed into competent cell with high efficiency, to get the library of alkane producing module. Because of the limitation of time, the size of the library is just 20,000. Just as Figure 4 shows, every plate had about 3000 colonies, more than 90% colonies were amplified fragments with correct length (~2000bp) by colony PCR(Figure 5), and 8 plates like this had been made. Then, the colonies were washed out and got together and then whole plasmids were extracted from it to get the amplified plasmid library of alkane producing module. This library was later transformed into the <i>E. coli</i> BL21 containing Alk-Selector. |
| - | [[Image: | + | [[Image:TJU-Project-A4.jpg|thumb|300px|center|<b>Figure 4.</b> Plate colony count result of library construction strains, with about 3000 colonies on the plate.]] |
| - | [[Image: | + | [[Image:TJU-Project-A5.jpg|thumb|500px|center|<b>Figure 5.</b> Colony PCR result of library construction strains. More than 90% colonies were amplified fragments with correct length (~2000bp)]] |
<br> | <br> | ||
| - | = Process of Selection and | + | = Process of Selection and Analysis = |
<br> | <br> | ||
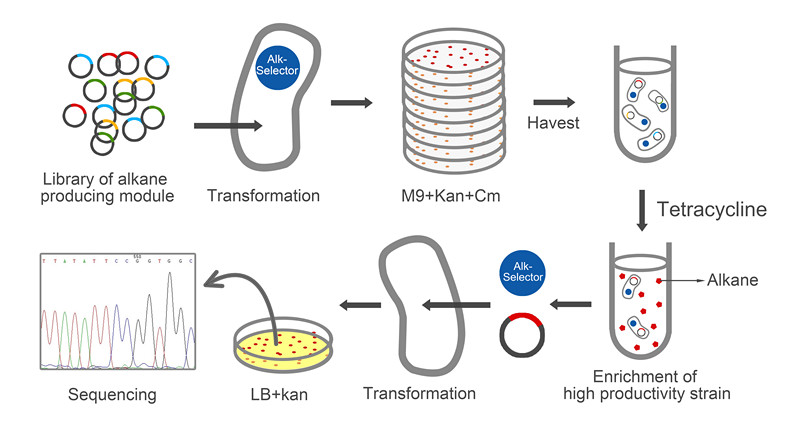
| - | The whole process of selection has been showed in Figure6. The amplified plasmid library was transformed to <i>E. coli</i> with Alk-Selector. To guarantee this library’s diversity, we think that the number of the strain which has successfully taken the alkane producing | + | The whole process of selection has been showed in Figure6. The amplified plasmid library was transformed to <i>E. coli</i> with Alk-Selector. To guarantee this library’s diversity, we think that the number of the strain which has successfully taken in the alkane producing module was at least 60,000. To verify this requirement, all the strains will be plated on the M9 solid medium, and the numbers of the colonies will be estimated. As Figure 6 showed that the number of 1/10 of all transformants is about 6500 which has satisfied the requirement. At the same time, the strains that didn’t take in the plasmid would eliminate under the pressure of antibiotic in this process. Then, all the colonies were washed out by M9 medium with antibiotics of proper concentration. The mix culture was inoculated into M9 medium with 12ug/ml tetracycline by 1:1000 for selection. For the control group, this culture was inoculated in the M9 medium without tetracycline. After fermentation for about 12 hours, <i>E. coli</i> began to grow very slowly, and after 36h, the OD600 of culture beyond 2.0 which was enough to analyze. |
| - | [[Image: | + | [[Image:TJU-Project-A6.jpg|thumb|600px|center|<b>Figure 6.</b> Selection and analysis process of the library with the help of Alk-Selector.]] |
| - | Because of the high copy number of Alk-Selector, directly | + | Because of the high copy number of Alk-Selector, directly separation of the colonies that survived during the selection process failed in the sequencing, so we extracted whole plasmid from the culture and retransformed to the E. coli without any module to discard the plasmid of Alk-Selector. The colonies will be picked up and sent for sequencing. After aligning the sequence, we can find the key animo acid which influences the function of the enzyme, and get the better module of alkane producing. |
<br> | <br> | ||
| Line 371: | Line 373: | ||
<br> | <br> | ||
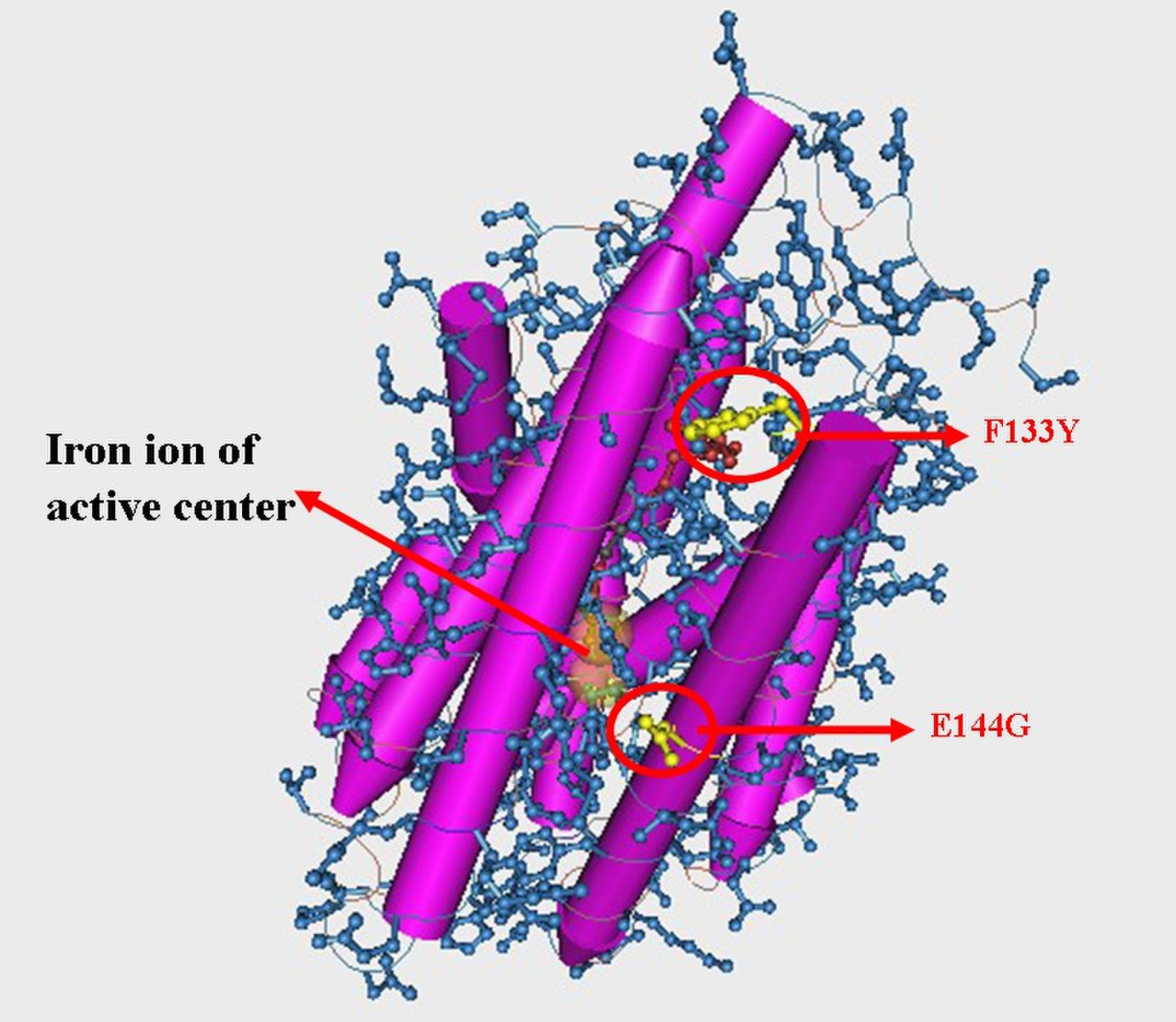
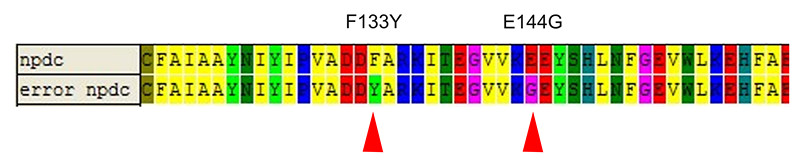
| - | We picked 50 colonies for sequencing. Limited by the size of the library, only 2 mutation has been found which is F133Y and E144G. Figure 7 and Figure 8 shows this result. About 80% strains contained both of the mutation (No strain from control experiment is enriched). Among two of the mutation, E144G is just nearby the key amino residue E145 which is responsible to stabilize the iron ions of the active center, so this mutation may | + | We picked 50 colonies for sequencing. Limited by the size of the library, only 2 mutation has been found which is F133Y and E144G. Figure 7 and Figure 8 shows this result. About 80% strains contained both of the mutation (No strain from control experiment is enriched). Among two of the mutation, E144G is just nearby the key amino residue E145 which is responsible to stabilize the iron ions of the active center, so this mutation may influence the activity of the enzyme. F133Y is far away to the active center, but may affect the entrance of the substrate to the pocket. Limited by the time, we didn’t test the function of the alkane producing module. |
| - | Anyway, we successfully | + | Anyway, we successfully finished our selection process from a library under the help of Alk-Selector. And we had proved that this favorite biobrick can work. |
| - | [[Image: | + | [[Image:TJU-PROJECT-A8.jpg|thumb|400px|center|<b>Figure 7.</b> 3D structure of enzyme NPDC with 2 mutation sites marked up: F133Y and E144G, and E144G is just nearby the key amino residue E145 related with active center. ]] |
| - | [[Image: | + | [[Image:TJU-Project-A7.jpg|thumb|650px|center|<b>Figure 8.</b> Protein sequences of NPDC and error NPDC, 2 mutation sites:F133Y and E144G have been found in survived NPDC.]] |
| Line 387: | Line 389: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/Introduction"><img src=" | + | <br/> |
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/Design&Construction"><img src=" | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Introduction"><img src="https://static.igem.org/mediawiki/2013/d/d3/TJU-project-banner01.jpg" width="120px" height="120px" align="left" alt="Introduction" title="Introduction"/></a> |
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/Characterization"><img src=" | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Design&Construction"><img src="https://static.igem.org/mediawiki/2013/a/a2/TJU-project-banner02.jpg" width="120px" height="120px" align="left" alt="Design & Construction" title="Design & Construction"/></a> |
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/ | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Characterization"><img src="https://static.igem.org/mediawiki/2013/9/9c/TJU-project-banner03.jpg" width="120px" height="120px" align="left" alt="Characterization" title="Characterization"/></a> |
| - | <a href="https://2013.igem.org/Team:Tianjin/Project/Futurework"><img src=" | + | <a href="https://2013.igem.org/Team:Tianjin/Project/Alk-Selector&DirectEvolution"><img src="https://static.igem.org/mediawiki/2013/9/98/TJU-project-banner04.jpg" width="120px" height="120px" align="left" alt="Alk-Selector And Direct Evolution" title="Alk-Selector And Direct Evolution"/></a> |
| + | <a href="https://2013.igem.org/Team:Tianjin/Project/Futurework"><img src="https://static.igem.org/mediawiki/2013/a/a2/TJU-project-banner05.jpg" width="120px" height="120px" align="left" alt="Futurework" title="Futurework"/></a> | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 03:07, 29 October 2013
After characterization of Alk-Sensor, we have proved that the AlkR-PalkM regulation system can recognize alkane produced by engineered strain and work well. With this property, we can select out the strains with high productivity of alkane easily according to the output reporter of AlkR-PalkM system. So in this session, we want to build a module called Alk-Selector to help us optimize the alkane biosynthetic pathway of engineered E. coli.
In our project, we choose NPDC gene, the key gene for alkane biosynthesis, as our target. We will randomly introduce mutation to the gene by error-prone PCR, and build a big library of the NPDC. Then, we will use our Alk-Selector to help us select out the best gene which can give E. coli a high productivity of alkane.
Contents |
Design and Construction of Alk-Selector
Firstly, we will design a new device Alk-Selector. Although Alk-Sensor can also be regarded as a selector, we don’t want to select just by our eyes which is a tough work. Instead, the special function of antibiotic resistant gene will help us select out the gene with better property automatically in the condition of antibiotic pressure.
So based on the construction of Alk-Sensor, we replaced the RFP reporter with tetracycline resistant gene TetA which is a transporter to pump tetracycline outward the cell. Other components of the Alk-Selector are same to Alk-Sensor. The final construction of Alk-Selector and its working mechanism can be seen in Figure 1. The alkane biosynthesis module expresses NPDC and AAR to produce alkane in E. coli, and then the alkane can activate the expression of tetA through AlkR-PalkM system, and the transporter TetA will give tetracycline resistance to E. coli and make it survive in the environment with tetracycline.
Characterization of Alk-Selector
To verify that Alk-Selector will generate more tetracycline resistance when more alkanes are produced, we cotransformed the Alk-Selector and an alkane producing module to the E. coli BL21. The control strain just had Alk-Selector. These two strains were cultured in M9 medium with 12ug/ml tetracycline. After 48h, we can find the growth of strain with alkane producing module was higher than the control just as Figure 2 showed.
It proved that our Alk-Selector can work.
Library Construction
For library construction, we reconstructed the alkane producing module of low productivity by adding restriction site BamHI and SacI at the two ends of gene NPDC for library construction. The workflow of the library construction is showed in Figure 3.
We amplified NPDC by error-prone PCR, and ligand on the plasmid. The product of ligation is transformed into competent cell with high efficiency, to get the library of alkane producing module. Because of the limitation of time, the size of the library is just 20,000. Just as Figure 4 shows, every plate had about 3000 colonies, more than 90% colonies were amplified fragments with correct length (~2000bp) by colony PCR(Figure 5), and 8 plates like this had been made. Then, the colonies were washed out and got together and then whole plasmids were extracted from it to get the amplified plasmid library of alkane producing module. This library was later transformed into the E. coli BL21 containing Alk-Selector.
Process of Selection and Analysis
The whole process of selection has been showed in Figure6. The amplified plasmid library was transformed to E. coli with Alk-Selector. To guarantee this library’s diversity, we think that the number of the strain which has successfully taken in the alkane producing module was at least 60,000. To verify this requirement, all the strains will be plated on the M9 solid medium, and the numbers of the colonies will be estimated. As Figure 6 showed that the number of 1/10 of all transformants is about 6500 which has satisfied the requirement. At the same time, the strains that didn’t take in the plasmid would eliminate under the pressure of antibiotic in this process. Then, all the colonies were washed out by M9 medium with antibiotics of proper concentration. The mix culture was inoculated into M9 medium with 12ug/ml tetracycline by 1:1000 for selection. For the control group, this culture was inoculated in the M9 medium without tetracycline. After fermentation for about 12 hours, E. coli began to grow very slowly, and after 36h, the OD600 of culture beyond 2.0 which was enough to analyze.
Because of the high copy number of Alk-Selector, directly separation of the colonies that survived during the selection process failed in the sequencing, so we extracted whole plasmid from the culture and retransformed to the E. coli without any module to discard the plasmid of Alk-Selector. The colonies will be picked up and sent for sequencing. After aligning the sequence, we can find the key animo acid which influences the function of the enzyme, and get the better module of alkane producing.
Result
We picked 50 colonies for sequencing. Limited by the size of the library, only 2 mutation has been found which is F133Y and E144G. Figure 7 and Figure 8 shows this result. About 80% strains contained both of the mutation (No strain from control experiment is enriched). Among two of the mutation, E144G is just nearby the key amino residue E145 which is responsible to stabilize the iron ions of the active center, so this mutation may influence the activity of the enzyme. F133Y is far away to the active center, but may affect the entrance of the substrate to the pocket. Limited by the time, we didn’t test the function of the alkane producing module.
Anyway, we successfully finished our selection process from a library under the help of Alk-Selector. And we had proved that this favorite biobrick can work.
 "
"