Team:Tianjin/Project/Design&Construction
From 2013.igem.org
(→b. what cause the leakage (from modeling information)) |
(→b. what's the influence of leakage? (from modeling information)) |
||
| (3 intermediate revisions not shown) | |||
| Line 329: | Line 329: | ||
<br> | <br> | ||
| - | == a. | + | == a. What cause the leakage?== |
| - | To find | + | To find the source fo the problem, we made a mathematical analysis to get the equation of leakage. (see more details on modeling part) From the equation, we found that the strength of PalkM, and the expression level of AlkR. To find the key of the leakage, we test the output of PalkM-RFP at the conditions with or without AlkR. The result shows in Fig 3, the strain with PalM-RFP and AlkR turned red, and the strain without AlkR is not red and no significant difference to BL21 which is without the gene of RFP. This test proved that the strength of PalkM is very small, and AlkR is the main source of the leakage. |
| - | ==b. | + | ==b.What's the influence of leakage? (from modeling information)== |
| - | + | To understand the influence of the leakage which is caused by AlkR, we modeled the relationship between input and output at different conditions with different expression level of AlkR. The result showed in Fig 4. , From the figure, we found that the dynamic range is negative related to the expression of AlkR, when the expression of AlkR is very high(just like the orange one), the output of the sensor will hardly change, the function of the sensor will be severely influenced. So it is important to reduce the expression of AlkR. | |
| - | + | ||
| - | + | ||
| - | + | ||
[[Image:TJU-project-F.png|center]] | [[Image:TJU-project-F.png|center]] | ||
Latest revision as of 03:01, 29 October 2013
Contents |
First Trail
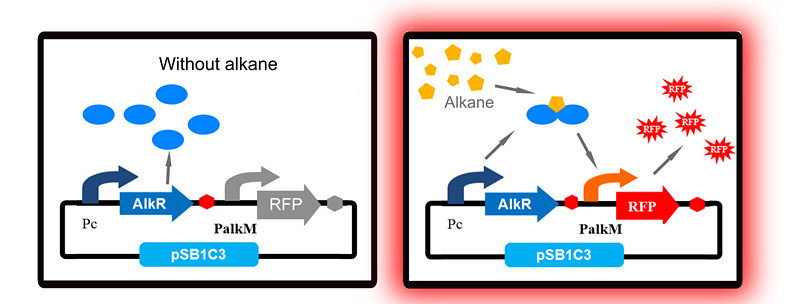
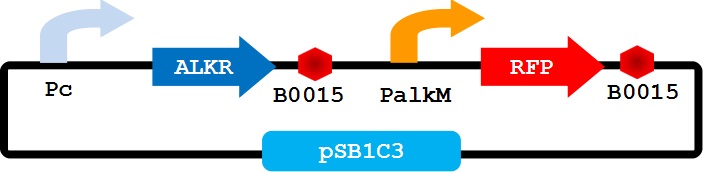
To apply the special alkane sensing system AlkR-PalkM of Acinetobacter baylyi ADP1 in E. coli, we built a gene circuit as Figure 1 showed. We used a constitutive promoter to control the expression of regulator AlkR, and RFP is controlled by PalkM. Without alkanes, the PalkM will not work, RFP will not be expressed. In the presence of alkanes, AlkR dimer will bind to the alkane molecules to form a complex, and the complex can turn on the expression of RFP which is controlled by PalkM. Then red fluorescence can be detected by naked eyes and measured by fluorescence spectrophotometer easily. Considering that another module may be put in E. coli to produce alkanes, we put Pc-AlkR-tt and PalkM-RFP-tt into one plasmid to form our Alk-Sensor.
Unfortunately, our first trial has a big problem: even in the absence of alkanes, strains with our Alk-Sensor turn red, which can be detected by naked eyes, as Fig 2 shows. The leakage of our Alk-Sensor is very high. This will influence alkane detection by naked eyes, so the Alk-Sensor need to be improved.
Analysis
a. What cause the leakage?
To find the source fo the problem, we made a mathematical analysis to get the equation of leakage. (see more details on modeling part) From the equation, we found that the strength of PalkM, and the expression level of AlkR. To find the key of the leakage, we test the output of PalkM-RFP at the conditions with or without AlkR. The result shows in Fig 3, the strain with PalM-RFP and AlkR turned red, and the strain without AlkR is not red and no significant difference to BL21 which is without the gene of RFP. This test proved that the strength of PalkM is very small, and AlkR is the main source of the leakage.
b.What's the influence of leakage? (from modeling information)
To understand the influence of the leakage which is caused by AlkR, we modeled the relationship between input and output at different conditions with different expression level of AlkR. The result showed in Fig 4. , From the figure, we found that the dynamic range is negative related to the expression of AlkR, when the expression of AlkR is very high(just like the orange one), the output of the sensor will hardly change, the function of the sensor will be severely influenced. So it is important to reduce the expression of AlkR.
Besides the high expression of AlkR, the low activity of the terminator downstream of the AlkR is also the source of leakage. In this condition, the transcription of constitutive promoter Pc may turn on the expression of RFP.
Summary of the leakage of RFP:
a)Promoter alkM has some leakage, to be more specific, it binds to RNA polymerase without undergoing a conformation change and activates the transcription.
b)Protein alkR or dimerized alkR could induce promoter alkM without alkane molecules.
c)The terminator downstream of AlkR is weak,and the constitutive promoter upstream of AlkR may influence the expression of RFP.
Reconstruction
To improve the function of the Alk-Sensor and decrease the leakage, we reconstructed our device.
We used a weaker promoter to control the expression of AlkR. And then, to reduce the interference of two transcription units, we used a stronger terminator to the downstream of two transcription units.
At last, we switched the position of the two transcription unit, to guarantee that PalkM-RFP isn’t interferenced by constitutive promoter. The new reconstructed Alk-Sensor shows in Figure 5.
Through another blank test we found that the reconstruction could decrease AlkSensor’s leakage significantly. After optimization, the percentage of bacterial colonies that are red enough to be distinguished by naked eyes decreased from 95% to 15%.
At last, we used full dose octane to induce the two sensors. After a certain period of cultivation, the relative fluorescent intensity was measured. The results showed that after reconstruction the leakage decreased by 50%, and the dynamic range increased by 5 folds, which means that the optimized AlkSensor has stronger ability to distinguish different inputs. ( showed in Figure 6)
So we choose this construction as our final design for Alk-Sensor and one of our favorite biobricks.
 "
"