26/07/13
From 2013.igem.org
(Difference between revisions)
AliceHaworth (Talk | contribs) (→Restriction enzyme digest of the DNA samples) |
|||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | # | + | <div style="font-family:arial;padding:5px;border-radius:5px;border:5px solid #FF2800;"> |
| - | + | {| style="color:#87EA00;background-color:#FFFFFF;" cellpadding="2" cellspacing="2" border="0" bordercolor="#000000" width="100%" align="center" | |
| - | + | !align="center"|[[Team:Leicester|Home]] | |
| - | + | !align="center"|[[Team:Leicester/Team|Team]] | |
| - | + | !align="center"|[https://igem.org/Team.cgi?year=2013&team_name=Leicester Official Team Profile] | |
| - | + | !align="center"|[[Team:Leicester/Project|Project]] | |
| - | + | !align="center"|[[Team:Leicester/Parts|Parts Submitted to the Registry]] | |
| - | + | !align="center"|[[Team:Leicester/Modeling|Modeling]] | |
| + | !align="center"|[[Team:Leicester/Notebook|Notebook]] | ||
| + | !align="center"|[[Team:Leicester/Safety|Safety]] | ||
| + | !align="center"|[[Team:Leicester/Attributions|Attributions]] | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | |||
| + | {|border=1 | ||
| + | |Sample||Plasmid Volume(ul)||DNA concentration (ng/ul) | ||
| + | |- | ||
| + | |1.1||40||131.8 | ||
| + | |- | ||
| + | |1.2||40||77.1 | ||
| + | |- | ||
| + | |1.3||46||56.8 | ||
| + | |- | ||
| + | |1.4||39||109 | ||
| + | |- | ||
| + | |1.5||41||129.1 | ||
| + | |- | ||
| + | |1.6||32||93.3 | ||
| + | |} | ||
| + | |||
*measured DNA concentration using nanodrop | *measured DNA concentration using nanodrop | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
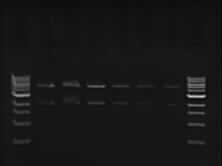
| - | + | ==Restriction enzyme digest of the DNA samples== | |
**enzymes: XbaI, PstI | **enzymes: XbaI, PstI | ||
**master mix: | **master mix: | ||
| Line 28: | Line 44: | ||
*To calculate it we divided 200ng by the concentration of each sample | *To calculate it we divided 200ng by the concentration of each sample | ||
*The rest of the volume was made up with water to give a total of 5ul. | *The rest of the volume was made up with water to give a total of 5ul. | ||
| - | *The samples were then incubated in a water bath at 37C | + | *The samples were then incubated in a water bath at 37C for an hour |
| - | + | ==Agorose Gel preparation== | |
***20ml of 5xTBE | ***20ml of 5xTBE | ||
***80ml of water | ***80ml of water | ||
Latest revision as of 14:15, 22 August 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
| Sample | Plasmid Volume(ul) | DNA concentration (ng/ul) |
| 1.1 | 40 | 131.8 |
| 1.2 | 40 | 77.1 |
| 1.3 | 46 | 56.8 |
| 1.4 | 39 | 109 |
| 1.5 | 41 | 129.1 |
| 1.6 | 32 | 93.3 |
- measured DNA concentration using nanodrop
Restriction enzyme digest of the DNA samples
- enzymes: XbaI, PstI
- master mix:
- Buffer - 14ul
- H2O - 88.2ul
- XbaI - 1.4ul
- PstI - 1.4ul
- Total volume - 105ul
- Each sample contained 5ul of DNA and 15ul of the master mix.
- The total amount of DNA in each sample was 200ng
- To calculate it we divided 200ng by the concentration of each sample
- The rest of the volume was made up with water to give a total of 5ul.
- The samples were then incubated in a water bath at 37C for an hour
Agorose Gel preparation
- 20ml of 5xTBE
- 80ml of water
- 0.8g of agarose
- Boil for 3 minutes, 1 minute at a time.
- Let it cool and add 5ul of ethidium bromide.
- Pour and let it set for 30 mins.
- Adding dye and marker
- Used orange g loading dye.
- 2ul to each sample
- Used 1kb ladder, 5ul per 1 marker well.
- Run the gel
- Gel picture
 "
"