Team:BostonU/Software
From 2013.igem.org
Tlozanoski (Talk | contribs) |
|||
| (15 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<br><br> | <br><br> | ||
<h2>Software Notebook</h2> | <h2>Software Notebook</h2> | ||
| - | <p>This notebook contains our work on various software projects, including Eugene and the | + | <p>This notebook contains our work on various software projects, including Eugene and the Datasheet App.</p> |
<br> | <br> | ||
<p> | <p> | ||
| Line 10: | Line 10: | ||
<strong>Subgoals:</strong> | <strong>Subgoals:</strong> | ||
<ul> | <ul> | ||
| - | <li>Use Eugene to | + | <li>Use Eugene to describe circuits</li> <strong> completed see <a href="https://2013.igem.org/Team:BostonU/Data">Data Collected </a></strong> |
| - | <li>Develop | + | <li>Develop datasheet app that will organize information for parts in the registry</li> |
<ul> | <ul> | ||
| + | <li>Work with Purdue to refine Boston U 2012 Datasheet draft </li><strong> completed </strong> | ||
| + | <li>Build functioning program to with intuitive user interface with comprehensive data fields</li><strong> preliminary design completed </strong> | ||
| + | <li>Build program to parse iGEM Parts Registry and format into datasheet standared</li><strong> in progress </strong> | ||
<li>Make app compatible with Clotho 3.0</li> | <li>Make app compatible with Clotho 3.0</li> | ||
</ul> | </ul> | ||
| Line 23: | Line 26: | ||
<h8>Eugene - June 18, 2013</h8> | <h8>Eugene - June 18, 2013</h8> | ||
| - | <p>We met with Ernst and Diana from the Eugene team. We now have access to the newest Clotho releases through the SVN, and Ernst created a dynamic naming system in the Eugene language so users can use for loops to name parts that have consecutive numbering systems. We are working on | + | <p>We met with Ernst and Diana from the Eugene team. We now have access to the newest Clotho releases through the SVN, and Ernst created a dynamic naming system in the Eugene language so users can use for loops to name parts that have consecutive numbering systems. We are working on the Promoter Characterization Devices (PCDs) eugene file, and trying to make an inverter example. We have been also populating a database in Clotho with the parts and their sequences. This was started with a file Sonya had provided at the Eugene tutorial meeting, and we are filling out the sequences from the iGEM registry and from our lab members' sequencing files.</p> |
<br> | <br> | ||
| - | < | + | <h8>Datasheet - July 2, 2013</h8> |
| - | <p>Today we met with Swapnil, Evan, and Jenhan to discuss the | + | <p>Today we met with Swapnil, Evan, and Jenhan to discuss the datasheet app. We discussed what fields on the datasheet should be auto-generated, user inputted, or part of a drop down menu.</p> |
<center><img src="https://static.igem.org/mediawiki/2013/d/db/Datasheet_01.png"></center> | <center><img src="https://static.igem.org/mediawiki/2013/d/db/Datasheet_01.png"></center> | ||
| - | <p>The Purdue Biomakers responded to Thomas’s email. Thomas had sent them our | + | <p>The Purdue Biomakers responded to Thomas’s email. Thomas had sent them our datasheet research and initial plans, and we got a positive response from them. They want to potentially collaborate on the datasheet.</p> |
<p>Evan and Jenhan met with Stephanie from the computational portion of the CIDAR lab. We’re still unsure whether we want the app to pull data from Clotho or the ICE Registry, which is a new registry of parts at BU. Evan suggested that we send our table to Stephanie to see what she thinks would work with Clotho versus ICE.</p> | <p>Evan and Jenhan met with Stephanie from the computational portion of the CIDAR lab. We’re still unsure whether we want the app to pull data from Clotho or the ICE Registry, which is a new registry of parts at BU. Evan suggested that we send our table to Stephanie to see what she thinks would work with Clotho versus ICE.</p> | ||
<p><ul>Action Items: | <p><ul>Action Items: | ||
| - | <li> | + | <li>still need to choose between Clotho 3.0 and ICE</li> |
| - | <li> | + | <li>still need to decide who will be coding the Datasheet</li> |
| - | <li> | + | <li>continue the dialogue with Purdue to see what feedback we can get on the DataSheet</li></ul></p> |
<br> | <br> | ||
| - | < | + | <h8>Eugene - July 9, 2013</h8> |
<p>Weekly meeting with Ernst and Diana</p> | <p>Weekly meeting with Ernst and Diana</p> | ||
<p>We brought up some problems we were having with specifying fusion sites in Eugene: how an AE device must go before an EF, and an EF before an FG and so on. The format in which the parts are inputted into the Clotho 2.0 Database has annotations for the fusion sites. So each version of part (J23100_AB, J23100_EB, J23100_FB) must be inputted separately, but the rules must be able to pick which one can fit that particular transcriptional unit. Instead of digging into the sequence property of the parts, we decided to add two extra properties: FSLeft and FSRight, to help match up promoters and terminators in separate but consecutive transcriptional units.</p> | <p>We brought up some problems we were having with specifying fusion sites in Eugene: how an AE device must go before an EF, and an EF before an FG and so on. The format in which the parts are inputted into the Clotho 2.0 Database has annotations for the fusion sites. So each version of part (J23100_AB, J23100_EB, J23100_FB) must be inputted separately, but the rules must be able to pick which one can fit that particular transcriptional unit. Instead of digging into the sequence property of the parts, we decided to add two extra properties: FSLeft and FSRight, to help match up promoters and terminators in separate but consecutive transcriptional units.</p> | ||
<br> | <br> | ||
| - | < | + | <h8>Eugene - July 16, 2013</h8> |
<p>Weekly meeting with Ernst and Diana</p> | <p>Weekly meeting with Ernst and Diana</p> | ||
<p>Upon completing and reviewing the scripts we generated for the pRepLib and the Inverters, we found out that the Eugene Rule Engine is not quite able to handle the high volume of possibilites presented by the large libraries we were pulling from with several rules. Ernst has been working on the next version of Eugene which will incorporate a more efficient rule engine. When that is available, he will let us know to try our scripts again. Diana also has shown us some of the progress she has made on the Eugene documentation page. We were very pleased with the professional layout and easy to navigate sections.</p> | <p>Upon completing and reviewing the scripts we generated for the pRepLib and the Inverters, we found out that the Eugene Rule Engine is not quite able to handle the high volume of possibilites presented by the large libraries we were pulling from with several rules. Ernst has been working on the next version of Eugene which will incorporate a more efficient rule engine. When that is available, he will let us know to try our scripts again. Diana also has shown us some of the progress she has made on the Eugene documentation page. We were very pleased with the professional layout and easy to navigate sections.</p> | ||
<br> | <br> | ||
| - | < | + | <h8>Datasheet - July 24, 2013</h8> |
| - | <p>The | + | <p>The Datasheet team connected with the Purdue Biomakers via Google Hangout to talk about our Datasheet projects. They walked us through their Protocol form and Datasheet, which had a few new changes and additions to the one we sent them. We were excited to hear that they wanted to use our datasheet as the basic format for the project. Afterwards, we filled out a survey for them, outlining some of the things we would like to see changed. We also asked them to send us a screencast of the program so we could see the functionality of the program. Evan and Jenhan asked us to update our DataSheet model with any changes we liked that Purdue had made, and make any new reorganizations we thought necessary. We liked the protocol form idea, to help set up the datasheet template. We are looking forward to incorporating that part into our project. As of now, both teams are going to work on their own datasheet apps, mainly because a large goal of ours is to get it to connect to a Clotho database. We hope to compare and contrast both apps later in the summer, and again see what changes could be made to improve it further. We planned to meet again a week from today with Evan and Jenhan to show them another draft of the datasheet.</p> |
<br> | <br> | ||
| - | < | + | <h8>Datasheet - July 25, 2013</h8> |
| - | <p>The members of the iGEM met together to talk some more about the | + | <p>The members of the iGEM met together to talk some more about the Datasheet. We decided we wanted to separate the Datasheets into two types: Basic Parts and Composite Parts.</p> |
| - | <p>We mapped out what sort of information would be required for each type of | + | <p>We mapped out what sort of information would be required for each type of Datasheet, and how we might incorporate some of the ideas that Purdue presented to us. We liked the idea of a restriction map being available, but thought it might take up too much space on the page. We liked the idea of a singular sheet holding all the information for the part. We decided we would re-use the existing presentation of the sequence map already on the registry. It has a link to a plasmid map, like the one Purdue had on their version of the datasheet.</p> |
| - | <p>We also talked with Sonya about what some of the other protocols might be used for in the | + | <p>We also talked with Sonya about what some of the other protocols might be used for in the datasheet. For the time being, we want to set the standards for flow cytometry, since that will be the main demonstration of functionality that we will be using.</p> |
<p>(Due to some scheduling conflicts with a meeting with Wellesley to try out their latest Eugenie program, the meeting with Jenhan and Evan was moved to the following week: 8/7/2013</p> | <p>(Due to some scheduling conflicts with a meeting with Wellesley to try out their latest Eugenie program, the meeting with Jenhan and Evan was moved to the following week: 8/7/2013</p> | ||
<br> | <br> | ||
| - | < | + | <h8>Datasheet - August 8, 2013</h8> |
| - | <p>Working off of last year’s team’s template for the datasheet, the Purdue revisions, and the BU 2013 revisions, we made changes to the | + | <p>Working off of last year’s team’s template for the datasheet, the Purdue revisions, and the BU 2013 revisions, we made changes to the datasheet format and presented them to Evan and Jenhan. We decided that Pooja and Devina will be the primary programmers for the project. We also received a screencast from the Purdue Biomakers team! We discussed some of the advantages and disadvantages of the program. Thomas, who is from Indiana, is going to try to visit the Purdue team during a short break he is taking to go home. (Check out the the Summer Fun page to see who Thomas ran into at the Boston Logan Airport!)</p> |
<br> | <br> | ||
| - | < | + | <h8>Datasheet - August 13, 2013</h8> |
| - | <p>Thomas visited Purdue University in West Lafayette, Indiana to meet with Charlotte Hoo and Chris Thompson of the Biomaker’s iGEM team. The teams mutually agreed that BostonU would take leadership on the implementation of the web application/Clotho app for the creation of new | + | <p>Thomas visited Purdue University in West Lafayette, Indiana to meet with Charlotte Hoo and Chris Thompson of the Biomaker’s iGEM team. The teams mutually agreed that BostonU would take leadership on the implementation of the web application/Clotho app for the creation of new datasheets while Purdue would head up the distribution of the web app through the various connections they had made while seeking out preliminary feedback on early datasheet prototypes. Using the prototypes BU had as well as the various revisions other teams had suggested to Purdue, Thomas, Charlotte, and Chris reviewed the proposed changes and made a final draft of the datasheet in a MS Word document to show iGEM HQ later this week.</p> |
<p>While BU has a good handle on necessary information for flow cytometry data and induction curves, Purdue is going to try to put together a set of standard data fields for mass spectrometry and reach out to other teams they have worked with to develop some standard data fields for other protocols often used to characterize devices.</p> | <p>While BU has a good handle on necessary information for flow cytometry data and induction curves, Purdue is going to try to put together a set of standard data fields for mass spectrometry and reach out to other teams they have worked with to develop some standard data fields for other protocols often used to characterize devices.</p> | ||
<p>Meanwhile, Devina and Pooja, with the help of our mentors Jenhan and Evan, have gotten a start on the web app back at Boston University.</p> | <p>Meanwhile, Devina and Pooja, with the help of our mentors Jenhan and Evan, have gotten a start on the web app back at Boston University.</p> | ||
<br> | <br> | ||
| - | < | + | <h8>Eugene - August 13, 2013</h8> |
<p>The quorum sensing script has been written for Eugene with minimal issues. We had reviewed Diana’s updates on the new documentation page and were very happy with her progress.</p> | <p>The quorum sensing script has been written for Eugene with minimal issues. We had reviewed Diana’s updates on the new documentation page and were very happy with her progress.</p> | ||
<br> | <br> | ||
| - | < | + | <h8>Datasheet - August 15, 2013</h8> |
<p>Devina and Pooja have been reading up on JavaScript and Java in preparation for more work on the app. They met with Evan and Jenhan to go over the plan of action for the app.</p> | <p>Devina and Pooja have been reading up on JavaScript and Java in preparation for more work on the app. They met with Evan and Jenhan to go over the plan of action for the app.</p> | ||
| + | <br> | ||
| + | <h8>Datasheet - August 19, 2013</h8> | ||
| + | <p>Devina has been working with Jenhan this week to develop the client side of the Datasheet App.</p> | ||
| + | <br> | ||
| + | |||
| + | <h8>Eugene - August 20, 2013</h8> | ||
| + | <p>Weekly meeting with Ernst and Diana</p> | ||
| + | <p>Ernst has his new version of Eugene that can handle the large data sets. He demonstrated its use for an inverter script. Devina re-sent him the pRepLib script to be updated to the new version of Eugene syntax. Pooja sent him the QS script to be updated as well.</p> | ||
| + | <br> | ||
| + | |||
| + | <h8>Datasheet - August 26, 2013</h8> | ||
| + | <p>Pooja has been working with Evan to familiarize herself with Java and develop the server side of the App.</p> | ||
| + | <br> | ||
| + | |||
| + | <h8>Datasheet - September 16, 2013</h8> | ||
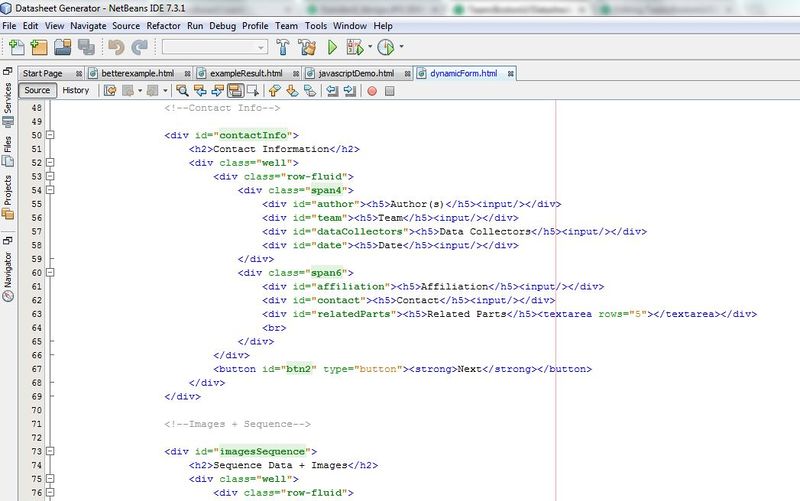
| + | <p>Devina is writing code to generate the user interface. Here is a sample:</p> | ||
| + | <p><center><img src="https://static.igem.org/mediawiki/2013/thumb/c/c9/Code.JPG/800px-Code.JPG" width="600px"></center></p> | ||
| + | <br> | ||
| + | |||
| + | <h8>Datasheet - September 19, 2013</h8> | ||
| + | <p>Pooja is working on some back-end server side code. Here is a sample:</p> | ||
| + | <p><center><img src="https://static.igem.org/mediawiki/2013/8/86/Backendcode.png" width="600px"></center></p> | ||
| + | <br> | ||
</html> | </html> | ||
Latest revision as of 03:28, 28 September 2013
Software Notebook
This notebook contains our work on various software projects, including Eugene and the Datasheet App.
Subgoals:
- Use Eugene to describe circuits completed see Data Collected
- Develop datasheet app that will organize information for parts in the registry
- Work with Purdue to refine Boston U 2012 Datasheet draft completed
- Build functioning program to with intuitive user interface with comprehensive data fields preliminary design completed
- Build program to parse iGEM Parts Registry and format into datasheet standared in progress
- Make app compatible with Clotho 3.0
We met with Ernst and Diana from the Eugene team. We now have access to the newest Clotho releases through the SVN, and Ernst created a dynamic naming system in the Eugene language so users can use for loops to name parts that have consecutive numbering systems. We are working on the Promoter Characterization Devices (PCDs) eugene file, and trying to make an inverter example. We have been also populating a database in Clotho with the parts and their sequences. This was started with a file Sonya had provided at the Eugene tutorial meeting, and we are filling out the sequences from the iGEM registry and from our lab members' sequencing files.
Today we met with Swapnil, Evan, and Jenhan to discuss the datasheet app. We discussed what fields on the datasheet should be auto-generated, user inputted, or part of a drop down menu.

The Purdue Biomakers responded to Thomas’s email. Thomas had sent them our datasheet research and initial plans, and we got a positive response from them. They want to potentially collaborate on the datasheet.
Evan and Jenhan met with Stephanie from the computational portion of the CIDAR lab. We’re still unsure whether we want the app to pull data from Clotho or the ICE Registry, which is a new registry of parts at BU. Evan suggested that we send our table to Stephanie to see what she thinks would work with Clotho versus ICE.
- Action Items:
- still need to choose between Clotho 3.0 and ICE
- still need to decide who will be coding the Datasheet
- continue the dialogue with Purdue to see what feedback we can get on the DataSheet
Weekly meeting with Ernst and Diana
We brought up some problems we were having with specifying fusion sites in Eugene: how an AE device must go before an EF, and an EF before an FG and so on. The format in which the parts are inputted into the Clotho 2.0 Database has annotations for the fusion sites. So each version of part (J23100_AB, J23100_EB, J23100_FB) must be inputted separately, but the rules must be able to pick which one can fit that particular transcriptional unit. Instead of digging into the sequence property of the parts, we decided to add two extra properties: FSLeft and FSRight, to help match up promoters and terminators in separate but consecutive transcriptional units.
Weekly meeting with Ernst and Diana
Upon completing and reviewing the scripts we generated for the pRepLib and the Inverters, we found out that the Eugene Rule Engine is not quite able to handle the high volume of possibilites presented by the large libraries we were pulling from with several rules. Ernst has been working on the next version of Eugene which will incorporate a more efficient rule engine. When that is available, he will let us know to try our scripts again. Diana also has shown us some of the progress she has made on the Eugene documentation page. We were very pleased with the professional layout and easy to navigate sections.
The Datasheet team connected with the Purdue Biomakers via Google Hangout to talk about our Datasheet projects. They walked us through their Protocol form and Datasheet, which had a few new changes and additions to the one we sent them. We were excited to hear that they wanted to use our datasheet as the basic format for the project. Afterwards, we filled out a survey for them, outlining some of the things we would like to see changed. We also asked them to send us a screencast of the program so we could see the functionality of the program. Evan and Jenhan asked us to update our DataSheet model with any changes we liked that Purdue had made, and make any new reorganizations we thought necessary. We liked the protocol form idea, to help set up the datasheet template. We are looking forward to incorporating that part into our project. As of now, both teams are going to work on their own datasheet apps, mainly because a large goal of ours is to get it to connect to a Clotho database. We hope to compare and contrast both apps later in the summer, and again see what changes could be made to improve it further. We planned to meet again a week from today with Evan and Jenhan to show them another draft of the datasheet.
The members of the iGEM met together to talk some more about the Datasheet. We decided we wanted to separate the Datasheets into two types: Basic Parts and Composite Parts.
We mapped out what sort of information would be required for each type of Datasheet, and how we might incorporate some of the ideas that Purdue presented to us. We liked the idea of a restriction map being available, but thought it might take up too much space on the page. We liked the idea of a singular sheet holding all the information for the part. We decided we would re-use the existing presentation of the sequence map already on the registry. It has a link to a plasmid map, like the one Purdue had on their version of the datasheet.
We also talked with Sonya about what some of the other protocols might be used for in the datasheet. For the time being, we want to set the standards for flow cytometry, since that will be the main demonstration of functionality that we will be using.
(Due to some scheduling conflicts with a meeting with Wellesley to try out their latest Eugenie program, the meeting with Jenhan and Evan was moved to the following week: 8/7/2013
Working off of last year’s team’s template for the datasheet, the Purdue revisions, and the BU 2013 revisions, we made changes to the datasheet format and presented them to Evan and Jenhan. We decided that Pooja and Devina will be the primary programmers for the project. We also received a screencast from the Purdue Biomakers team! We discussed some of the advantages and disadvantages of the program. Thomas, who is from Indiana, is going to try to visit the Purdue team during a short break he is taking to go home. (Check out the the Summer Fun page to see who Thomas ran into at the Boston Logan Airport!)
Thomas visited Purdue University in West Lafayette, Indiana to meet with Charlotte Hoo and Chris Thompson of the Biomaker’s iGEM team. The teams mutually agreed that BostonU would take leadership on the implementation of the web application/Clotho app for the creation of new datasheets while Purdue would head up the distribution of the web app through the various connections they had made while seeking out preliminary feedback on early datasheet prototypes. Using the prototypes BU had as well as the various revisions other teams had suggested to Purdue, Thomas, Charlotte, and Chris reviewed the proposed changes and made a final draft of the datasheet in a MS Word document to show iGEM HQ later this week.
While BU has a good handle on necessary information for flow cytometry data and induction curves, Purdue is going to try to put together a set of standard data fields for mass spectrometry and reach out to other teams they have worked with to develop some standard data fields for other protocols often used to characterize devices.
Meanwhile, Devina and Pooja, with the help of our mentors Jenhan and Evan, have gotten a start on the web app back at Boston University.
The quorum sensing script has been written for Eugene with minimal issues. We had reviewed Diana’s updates on the new documentation page and were very happy with her progress.
Devina and Pooja have been reading up on JavaScript and Java in preparation for more work on the app. They met with Evan and Jenhan to go over the plan of action for the app.
Devina has been working with Jenhan this week to develop the client side of the Datasheet App.
Weekly meeting with Ernst and Diana
Ernst has his new version of Eugene that can handle the large data sets. He demonstrated its use for an inverter script. Devina re-sent him the pRepLib script to be updated to the new version of Eugene syntax. Pooja sent him the QS script to be updated as well.
Pooja has been working with Evan to familiarize herself with Java and develop the server side of the App.
Devina is writing code to generate the user interface. Here is a sample:
Pooja is working on some back-end server side code. Here is a sample:

 "
"

