22/08/13
From 2013.igem.org
(Difference between revisions)
AliceHaworth (Talk | contribs) (→Measuring the concentrations of isolated P. putida DNA from the previous day) |
AliceHaworth (Talk | contribs) |
||
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <div style="font-family:arial;padding:5px;border-radius:5px;border:5px solid #FF2800;"> | ||
| + | {| style="color:#87EA00;background-color:#FFFFFF;" cellpadding="2" cellspacing="2" border="0" bordercolor="#000000" width="100%" align="center" | ||
| + | !align="center"|[[Team:Leicester|Home]] | ||
| + | !align="center"|[[Team:Leicester/Team|Team]] | ||
| + | !align="center"|[https://igem.org/Team.cgi?year=2013&team_name=Leicester Official Team Profile] | ||
| + | !align="center"|[[Team:Leicester/Project|Project]] | ||
| + | !align="center"|[[Team:Leicester/Parts|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Leicester/Modeling|Modeling]] | ||
| + | !align="center"|[[Team:Leicester/Notebook|Notebook]] | ||
| + | !align="center"|[[Team:Leicester/Safety|Safety]] | ||
| + | !align="center"|[[Team:Leicester/Attributions|Attributions]] | ||
| + | |} | ||
| + | </div> | ||
==Measuring the concentrations of isolated ''P. putida'' DNA from the previous day== | ==Measuring the concentrations of isolated ''P. putida'' DNA from the previous day== | ||
*Measuring concentration using nanodrop | *Measuring concentration using nanodrop | ||
*Blanking against buffer | *Blanking against buffer | ||
| - | {| | + | {|border=1 |
|Sample nr.||Concentration ng/ul||260/280||260/230 | |Sample nr.||Concentration ng/ul||260/280||260/230 | ||
|- | |- | ||
| Line 9: | Line 22: | ||
|4||30.6||1.8||1.29 | |4||30.6||1.8||1.29 | ||
|- | |- | ||
| - | |6||42.3||1.81|1.05 | + | |6||42.3||1.81||1.05 |
|- | |- | ||
|8||35.6||1.85||1.26 | |8||35.6||1.85||1.26 | ||
| Line 21: | Line 34: | ||
|16||18.5||1.17||0.67 | |16||18.5||1.17||0.67 | ||
|} | |} | ||
| + | |||
| + | ==Preparing samples to run on 1% gel== | ||
| + | *500ng of each of the isolated ''P. putida'' DNA sample is run on gel | ||
| + | *Volumes are adjusted to 20ul per well accordingly (includes DNA, sterile H2O and OG) | ||
| + | *5ul of orange G dye is added to each sample | ||
| + | *5ul of 1kb ladders are used | ||
| + | *For sheared DNA, dilutions were loaded | ||
| + | **1ul of DNA was diluted in 9ul of 1xTE buffer to make up 10x dilution | ||
| + | **1ul from 10x dilution was put in 9ul of 1xTE buffer to make up 100x dilution | ||
| + | *Dilutions were nanodropped and the concentration for 100 fold dilution was 121 ng/ul | ||
| + | *200ng of herring sperm DNA was run on gel | ||
| + | *One lane was left for control, non-sheared DNA, also 100 fold dilution | ||
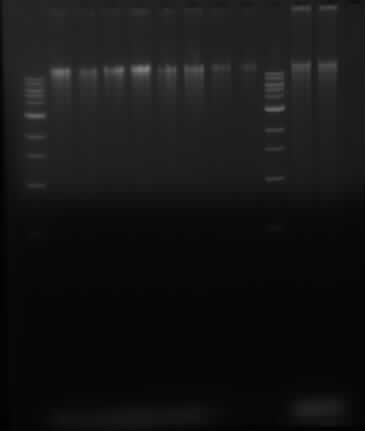
==Running gel of ''P. putida'' DNA and Herring sperm DNA== | ==Running gel of ''P. putida'' DNA and Herring sperm DNA== | ||
| Line 29: | Line 54: | ||
Samples in lanes goes as follows: 1 Kb ladder, 2, 4, 6, 8, 10, 12, 14, 16, 1 kb ladder, 100x dilution of sheared herring sperm DNA, control 100x diluted herring sperm DNA | Samples in lanes goes as follows: 1 Kb ladder, 2, 4, 6, 8, 10, 12, 14, 16, 1 kb ladder, 100x dilution of sheared herring sperm DNA, control 100x diluted herring sperm DNA | ||
| + | |||
| + | ==Running sonicated herring sperm DNA on 0.8% gel== | ||
| + | *3 lanes for running: | ||
| + | **Sheared and sonicated | ||
| + | **Unsheared and sonicated | ||
| + | **Unsheared and unsonicated | ||
| + | *5ul of 1kb marker is used | ||
| + | *The DNA samples are diluted 100 fold and 200ng of sample is run. | ||
| + | *Volume of sterile water is adjusted appropriately to make up 20ul of sample per well, including 5ul of orange G dye | ||
| + | [[File:igem_shear_son_220813.jpg]] | ||
| + | *Not visible on the computer file, but picture shows a very slight decrease in sheared/sonificated and unsheared/sonicated samples vs the unsheared/unsonificated control | ||
| + | *The difference is not significant, but the viscosity has changed when mechanically spreading the DNA on polystyrene, though it is not that evident on the gel. | ||
| + | |||
| + | ==Making up a sheared/sonicated dilution to leave on bench overnight== | ||
| + | *500ul of sheared/sonicated DNA | ||
| + | *500ul of 1xTE | ||
| + | *Mix | ||
| + | *Leave on bench overnight | ||
| + | |||
| + | ==Incubating sheared/sonicated dilution at 37C on a shaking rack== | ||
| + | *10ul of 1xTE added to 2ul of sheared/sonicated DNA | ||
| + | *Mix | ||
| + | *Incubated overnight at 37C on a shaking rack | ||
Latest revision as of 13:57, 23 August 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Measuring the concentrations of isolated P. putida DNA from the previous day
- Measuring concentration using nanodrop
- Blanking against buffer
| Sample nr. | Concentration ng/ul | 260/280 | 260/230 |
| 2 | 44 | 1.8 | 1.15 |
| 4 | 30.6 | 1.8 | 1.29 |
| 6 | 42.3 | 1.81 | 1.05 |
| 8 | 35.6 | 1.85 | 1.26 |
| 10 | 38.2 | 1.84 | 1.28 |
| 12 | 40.6 | 1.83 | 1.45 |
| 14 | 39.9 | 1.88 | 1.47 |
| 16 | 18.5 | 1.17 | 0.67 |
Preparing samples to run on 1% gel
- 500ng of each of the isolated P. putida DNA sample is run on gel
- Volumes are adjusted to 20ul per well accordingly (includes DNA, sterile H2O and OG)
- 5ul of orange G dye is added to each sample
- 5ul of 1kb ladders are used
- For sheared DNA, dilutions were loaded
- 1ul of DNA was diluted in 9ul of 1xTE buffer to make up 10x dilution
- 1ul from 10x dilution was put in 9ul of 1xTE buffer to make up 100x dilution
- Dilutions were nanodropped and the concentration for 100 fold dilution was 121 ng/ul
- 200ng of herring sperm DNA was run on gel
- One lane was left for control, non-sheared DNA, also 100 fold dilution
Running gel of P. putida DNA and Herring sperm DNA
Samples in lanes goes as follows: 1 Kb ladder, 2, 4, 6, 8, 10, 12, 14, 16, 1 kb ladder, 100x dilution of sheared herring sperm DNA, control 100x diluted herring sperm DNA
Running sonicated herring sperm DNA on 0.8% gel
- 3 lanes for running:
- Sheared and sonicated
- Unsheared and sonicated
- Unsheared and unsonicated
- 5ul of 1kb marker is used
- The DNA samples are diluted 100 fold and 200ng of sample is run.
- Volume of sterile water is adjusted appropriately to make up 20ul of sample per well, including 5ul of orange G dye
- Not visible on the computer file, but picture shows a very slight decrease in sheared/sonificated and unsheared/sonicated samples vs the unsheared/unsonificated control
- The difference is not significant, but the viscosity has changed when mechanically spreading the DNA on polystyrene, though it is not that evident on the gel.
Making up a sheared/sonicated dilution to leave on bench overnight
- 500ul of sheared/sonicated DNA
- 500ul of 1xTE
- Mix
- Leave on bench overnight
Incubating sheared/sonicated dilution at 37C on a shaking rack
- 10ul of 1xTE added to 2ul of sheared/sonicated DNA
- Mix
- Incubated overnight at 37C on a shaking rack
 "
"