Team:Tsinghua-E/Notebook
From 2013.igem.org
(→Week7(8.6-8.13)) |
(→Week7(8.6-8.13)) |
||
| Line 72: | Line 72: | ||
== '''Week7(8.6-8.13)''' == | == '''Week7(8.6-8.13)''' == | ||
[[File:Note0805.png|300px|thumb|left]][[File:Note0806.png|300px|thumb|right|Figure. 9 sweet pressure part construction with pBAD as promoter]] | [[File:Note0805.png|300px|thumb|left]][[File:Note0806.png|300px|thumb|right|Figure. 9 sweet pressure part construction with pBAD as promoter]] | ||
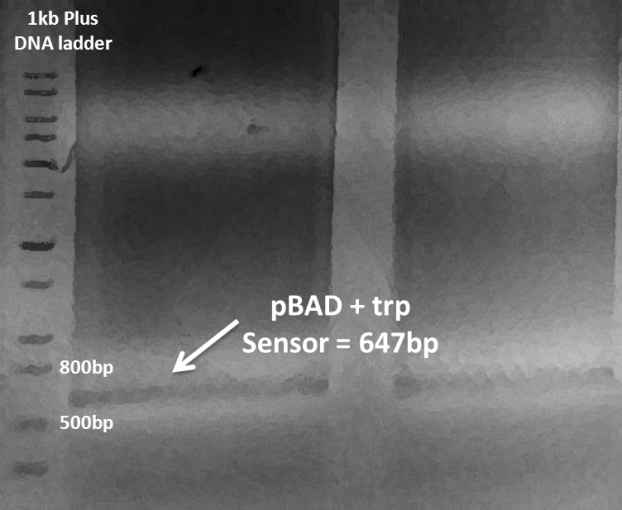
| - | Simultaneously, because of the failure of our sweet part to have desired growth dependence on tryptophan measured in the last week, we redesigned our circuit and replaced tac promoter in pTrc99A with more strict araBAD promoter (pBAD). At the same time, we designed random RBS library to control the expression level of malQ. In this way, a proper maltase expression level could be selected and expected to avoid the overexpression problem. The pBAD sequence was derived from (pRedET (amp),produced by Red^R/ ET^RRecombination). Hence, pBAD and tryptophan sensor were PCR amplified, gel-purified and assembled by overlap PCR. | + | Simultaneously, because of the failure of our sweet part to have desired growth dependence on tryptophan measured in the last week, we redesigned our circuit and replaced tac promoter in pTrc99A with more strict araBAD promoter (pBAD). At the same time, we designed random RBS library to control the expression level of malQ. In this way, a proper maltase expression level could be selected and expected to avoid the overexpression problem. The pBAD sequence was derived from (pRedET (amp),produced by Red^R/ ET^RRecombination). Hence, pBAD and tryptophan sensor were PCR amplified, gel-purified and assembled by overlap PCR. |
| - | + | ||
[[File:Part1I.png|420px|thumb|right|Figure. 9 sweet pressure part construction with pBAD as promoter]] | [[File:Part1I.png|420px|thumb|right|Figure. 9 sweet pressure part construction with pBAD as promoter]] | ||
| + | A 5’ end RBS random primer (malQ_RBS_library-F) for malQ was designed to produce the RBS library. pTrc99A plasmid was PCR amplified to remove tac promoter sequence and simultaneously linearized. These three gel-purified fragments were assembled by Takara infusion cloning kit. The plasmid with a random RBS library for malQ was named after pTrc99A_pBAD_trp sensor_random RBS_malQ. For the poor efficiency of the E. Coli JW3379 component cell, we firstly transformed the infusion product into E. Coli DH5a component cells and 120 single colonies were cultured and confirmed by PCR. After this step, we cocultured confirmed transformants in a 10mL LB medium and extracted the plasmid with the 〖E.Z.N.A.〗^TM plasmid mini kit I (produced by Omega Bio-tek). The plasmid was transformed into JW3379 and 48 single clones were picked. JW3379 chemically component cells were prepared by the following protocol. This library was named after sweet pressure part RBS library in this iGEM project. | ||
| + | |||
[[File:Note0807.png|420px|thumb|right|Figure. 9 sweet pressure part construction with pBAD as promoter]] | [[File:Note0807.png|420px|thumb|right|Figure. 9 sweet pressure part construction with pBAD as promoter]] | ||
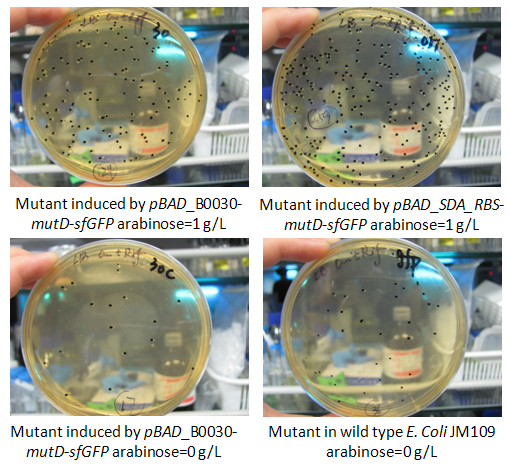
In this week, we also measured the mutation rate increase induced by mutD mutator. The basic protocol was provided as attachment “mutation rate measurement”. To be brief, strains E. Coli JM109 carrying vectors pBAD_B0030-mutD-sfGFP, pBAD_B0032-mutD-sfGFP and pBAD_SDA_RBS-mutD- sfGFP were cultured in LB medium and induced by 1g/L arabinose and the specific fluorescence of sfGFP (excitation 488nm, emission 512nm) (fluorescence to OD600) was measured. Thus, we made sure that pBAD_B0030-mutD-sfGFP, and pBAD_SDA_RBS-mutD- sfGFP have relatively higher mutD expression. Thus, only the mutation rate of pBAD_B0030-mutD-sfGFP, and pBAD_SDA_RBS-mutD- sfGFP were measured in the condition of arabinose induction by 1g/L and strains carrying pBAD_B0030-mutD-sfGFP without arabinose induction and wild type JM109 were set as negative control. We followed the protocol provided and found that pBAD_SDA_RBS-mutD- sfGFP could increase the genome mutation rate up to 10 times compared with negative control. The results were shown as the figure below. | In this week, we also measured the mutation rate increase induced by mutD mutator. The basic protocol was provided as attachment “mutation rate measurement”. To be brief, strains E. Coli JM109 carrying vectors pBAD_B0030-mutD-sfGFP, pBAD_B0032-mutD-sfGFP and pBAD_SDA_RBS-mutD- sfGFP were cultured in LB medium and induced by 1g/L arabinose and the specific fluorescence of sfGFP (excitation 488nm, emission 512nm) (fluorescence to OD600) was measured. Thus, we made sure that pBAD_B0030-mutD-sfGFP, and pBAD_SDA_RBS-mutD- sfGFP have relatively higher mutD expression. Thus, only the mutation rate of pBAD_B0030-mutD-sfGFP, and pBAD_SDA_RBS-mutD- sfGFP were measured in the condition of arabinose induction by 1g/L and strains carrying pBAD_B0030-mutD-sfGFP without arabinose induction and wild type JM109 were set as negative control. We followed the protocol provided and found that pBAD_SDA_RBS-mutD- sfGFP could increase the genome mutation rate up to 10 times compared with negative control. The results were shown as the figure below. | ||
Revision as of 17:30, 23 September 2013



Notice:Except for specially mentioned, all the following vectors were constructed by In-Fusion HD Cloning Kit (produced by Clontech). To be brief, every fragment of desired DNA was linearized by primers designed following the protocol provided by Takara infusion cloning guide to ensure accurately 15 base pair (bp) overlap homology with each adjacent fragments. The gel-purified fragments were mixed with 2uL infusion cloning kit Premix solution and ddH2O was used to adjust the mixture to 10uL. The amount of the added fragments was based on the Takara infusion cloning guide.
Contents |
Week1(6.23-6.30)
Literature was reviewed carefully and the corresponding gene candidates were confirmed. Some of them were submitted to be synthesized. Some of students were trained for basic molecule biology experiment operation.
Week2(7.1-7.7)
The primers needed in our project is designed and synthesized. Some chemicals were also purchased. Some of students were trained for basic molecule biology experiment operation and further some characterization experiment operation.
Week3(7.8-7.14)
We cloned dnaQ gene from E. Coli BL21(DE3) by PCR and utilized overlap extension PCR to introduce mutagenesis in dnaQ by the following protocol.
Firstly, we used E. Coli BL21 (DE3) strain as template to amplify dnaQ gene by PCR with mut-F and mut-R primers, respectively. After purification of PCR product (the 〖E.Z.N.A.〗^TM Gel Extraction Kit, produced by Omega Bio-tek), it was used as template to introduce point mutation by two parallel PCRs with the following two sets of primers: mut-L73W-F and mut-L73W-R (generating L73W); mut-A164V-F and mut-A164V-R (generating A164V). The three PCR products were gel-purified and combined by 5ng,respectively and assembled with mut-F and mut-R primers. The resulting overall product was T-cloned into Takara pMD-19 T-vector (produced by Takara BioTechnology (DALIAN)CO.,LTD.) and sequenced to confirm the sequence. The L73W and A164V mutant BL21 (DE3) dnaQ was named after mutD.
Maltose hydrolase gene malQ was cloned from E. Coli JW3995 (citation a malQ PCR is needed).
In parallel, we also constructed tryptophan sensor circuit. According to the report about the translating ribosome by nascent peptide based tryptophan degradation gene cluster regulation mechanism, we previously synthesized the corresponding tnaC coupling Rho sequence. This gene cluster was assembled with lacZ gene by overlap PCR and the gel-purified fragment was cloned between NcoI and HindIII sites of pTrc99A vector by restriction enzyme digest and T4 ligase induced ligation. All the information about primers was shown in “primer information”. This plasmid was named after pTrc99A_trp sensor_lacZ.
Week4(7.15-7.22)
In this week, we tried to clone mutD and sfGFP cluster downstream of araBAD promoter (pBAD). We amplified pSC101-Cm-pBAD and sfGFP fragment from plasmid AraC_pBAD_CI_OR222-sfGFP, which was kindly provided by Professor Ouyang Qi in Peking University. mutD was introduced by three different ribozyme binding site (RBS) sequence upstream and also overlap homology with primers. All the primers information was listed in the appendix, primer information. The three fragments were assembled by infusion cloning as described above. The partial sequence of RBS-mutD op-sfGFP was sequenced to confirm. The three constructed plasmids were named after pBAD_B0030-mutD-sfGFP, pBAD_B0032-mutD-sfGFP and pBAD_SDA_RBS-mutD-sfGFP, respectively.
In this week, we also tested our synthetic tryptophan sensor performance by galactosidase activity. Some protocol seemed not to work for some culture condition and measurement conditions. The final successful protocol was shown as below. Firstly, we picked up E. Coli JM109 carrying pTrc99A_trp sensor_lacZ single clone and cultured it in Luria-Bertani (LB) medium (Tryptone 10g/L, Yeast Extract 5g/L, NaCl 10g/L) with concentration of ampicillin at 50mg/L for overnight. On the next morning, the 600nm optical density (OD600) of the culture was measured to be 3.090. This seed was transferred into 2mL LB medium with the same concentration of ampicillin by a ratio of 5% in a 5mL 48 well culture plate (Huayang Chengxun Cor. Beijing China). After 1.6h, OD600 of every well reached about 0.6. Each well was also supplemented with 720uL fresh LB medium. Isopropylβ-D-1-Thiogalactopyranoside (IPTG) and tryptophan stock solution was also added into each well to the final concentration of 0.4mM and a gradient of 0.4, 0.5, 0.6, 0.8, 1, 2, 3mM, respectively. After 21h, the galactosidase activity was measured as described below.
Firstly, OD600 of the final culture was measured. Then mix 400uL culture with 2.8mL Z buffer (Na2HPO4.7H2O 16.1g/L, NaH2PO4.H2O 5.5g/L, KCl 0.75g/L, MgSO4.7H2O 0.25g/L, 2-thiol ethanol 2.7ml/L, adjusted pH to 7.0). Nextly, mix 2mL mixture with 250uL 1mg/mL MUG DMSO solution to react for 15min. Finally, 300uL 1M NaOH solution was added to quench the reaction and the fluorescence was measured with excitation and emission wavelength at 360 and 445 nm, respectively. The fluorescence intensity was normalized by culture OD600 and reaction time 15min as the activity of galactosidase (MUG units=F360/445/OD600/t). The results of the test were shown in Figure. 4.
Week5(7-23-7.29)
We constructed a plasmid expressing malQ responding to the concentration of tryptophan. Tryptophan sensor part and malQ were PCR amplified and gel-purified while pTrc99A was linearized between NcoI and PstI. Then the fragments were assembled by Takara infusion cloning kit. The plasmid was named pTrc99A_trp sensor_malQ. We firstly transformed the infusion product into DH5a component cell and 20 single colonies were cultured and confirmed by PCR. And then we extracted the plasmid from the confirmed transformants with the 〖E.Z.N.A.〗^TM plasmid mini kit I (produced by Omega Bio-tek) and transformed the plasmid into E. Coli JW3379.
After completing the construction, we tested whether the E.coli JW3379 transformed with pTrc99A_trp sensor_malQ have the growth dependence on tryptophan. We cultured the transformed E. Coli JW3379 in M9 media (100 mL Solution I(NaCl 0.5g/L,NH_4 Cl 1g/L ,Na_2 HPO_4∙〖12H〗_2 O 17.1g/L, KH_2 PO_4 3g/L),100 µL 1M MgSO_4,500µL 0.1M CaCl_2 ,4mL 20%(mass fraction) maltose) containing maltose as the single carbon source and added tryptophan to a concentration gratitude of 0 mM、1 mM、2 mM、3 mM、10 mM、20mM while adding the IPTG with OD600 reaching 0.6. The growth curve was shown in Figure.6.
Opposing to our expectation, the growth has a negative dependence on tryptophan. It may be caused by the little demand to malQ of the cell and thus the high expression level of malQ can become a burden to the growth of the cell. This problem might be more disturbing for tac promoter in original plasmid pTrc99A was not strict so the leakage effect was serious.
Week6(7.30-8.5)
To confirm our hypothesis about the tryptophan dependent malQ expression system, we tested the enzyme activity of malQ present in the cell 12 hours after adding IPTG. The enzyme activity is defined as the moles of maltose to glucose convertion per minute per OD600 at 37℃. And the glucose assay was conducted using the EnzyChromTM Glucose Assay Kit (produced by BioAssay Systems). We plotted the enzyme activity against the concentration of tryptophan and the result was shown as figure.7.
It was shown that when the concentration of tryptophan was below 10mM, the maltase activity increased with the level of tryptophan, which was the same as our expectation. However, when the concentration of tryptophan continued to increase, the enzyme activity started to decrease, showing the upper limit of tryptophan to be more or less 10mM.
In parallel, we constructed tryptophan sensor coupling tetA circuits which can provide selection pressure to tryptophan provider microorganisms by antibiotics resistance. Similar to mutD, three different RBS sequences (B0030, B0032, RBS derived from the upstream of wild type tnaA) were introduced upstream of tetA (cloned from pCM110) by PCR amplification. Plasmid pTrc99A_trp sensor_lacZ was also linearized by PCR to remove lacZ and at the same time introduced with tetA overlap homology. All the primers information was listed in the appendix, primer information. The partial functional part of this plasmid was sequenced. The three constructed plasmids were named after pTrc99A_trp sensor_B0030_tetA, pTrc99A_trp sensor_B0032_tetA and pTrc99A_trp sensor_B0030_tetA, respectively. In this iGEM project, they were named after bitter pressure part 30, 32 and ori, respectively.
Week7(8.6-8.13)
Simultaneously, because of the failure of our sweet part to have desired growth dependence on tryptophan measured in the last week, we redesigned our circuit and replaced tac promoter in pTrc99A with more strict araBAD promoter (pBAD). At the same time, we designed random RBS library to control the expression level of malQ. In this way, a proper maltase expression level could be selected and expected to avoid the overexpression problem. The pBAD sequence was derived from (pRedET (amp),produced by Red^R/ ET^RRecombination). Hence, pBAD and tryptophan sensor were PCR amplified, gel-purified and assembled by overlap PCR.
A 5’ end RBS random primer (malQ_RBS_library-F) for malQ was designed to produce the RBS library. pTrc99A plasmid was PCR amplified to remove tac promoter sequence and simultaneously linearized. These three gel-purified fragments were assembled by Takara infusion cloning kit. The plasmid with a random RBS library for malQ was named after pTrc99A_pBAD_trp sensor_random RBS_malQ. For the poor efficiency of the E. Coli JW3379 component cell, we firstly transformed the infusion product into E. Coli DH5a component cells and 120 single colonies were cultured and confirmed by PCR. After this step, we cocultured confirmed transformants in a 10mL LB medium and extracted the plasmid with the 〖E.Z.N.A.〗^TM plasmid mini kit I (produced by Omega Bio-tek). The plasmid was transformed into JW3379 and 48 single clones were picked. JW3379 chemically component cells were prepared by the following protocol. This library was named after sweet pressure part RBS library in this iGEM project.
In this week, we also measured the mutation rate increase induced by mutD mutator. The basic protocol was provided as attachment “mutation rate measurement”. To be brief, strains E. Coli JM109 carrying vectors pBAD_B0030-mutD-sfGFP, pBAD_B0032-mutD-sfGFP and pBAD_SDA_RBS-mutD- sfGFP were cultured in LB medium and induced by 1g/L arabinose and the specific fluorescence of sfGFP (excitation 488nm, emission 512nm) (fluorescence to OD600) was measured. Thus, we made sure that pBAD_B0030-mutD-sfGFP, and pBAD_SDA_RBS-mutD- sfGFP have relatively higher mutD expression. Thus, only the mutation rate of pBAD_B0030-mutD-sfGFP, and pBAD_SDA_RBS-mutD- sfGFP were measured in the condition of arabinose induction by 1g/L and strains carrying pBAD_B0030-mutD-sfGFP without arabinose induction and wild type JM109 were set as negative control. We followed the protocol provided and found that pBAD_SDA_RBS-mutD- sfGFP could increase the genome mutation rate up to 10 times compared with negative control. The results were shown as the figure below.
Week8
Week9
Week10
 "
"