Team:TU Darmstadt/safety/Labjournal
From 2013.igem.org
(Difference between revisions)
| Line 162: | Line 162: | ||
<font size="3" color="#F0F8FF" face="Arial regular"> | <font size="3" color="#F0F8FF" face="Arial regular"> | ||
| - | <p text-aligne:left style="margin-left:50px | + | <p text-aligne:left style="margin-left:50px; margin-right:50px"> |
| - | + | ||
Assembly of the gBlocks | Assembly of the gBlocks | ||
| Line 169: | Line 168: | ||
<Br> | <Br> | ||
| - | |||
| - | + | Based on the pDawn Plasmid we desinged the light induced kill switch and synthesized the construct as gBlock Fragments on IDT. We assembeled the 10 Fragments follow the Protocol: | |
| + | |||
<Br> | <Br> | ||
| + | |||
| + | <img alt="Assembly PCR with 10 gBlocks" src="/wiki/images/0/03/Assembly_pcr.jpg" width="100" height="200" align="right"></a> | ||
<ul style="margin-left:50px; margin-right:50px; text-align:justify; "> | <ul style="margin-left:50px; margin-right:50px; text-align:justify; "> | ||
| - | <li> | + | <li>Reconstitute the gBlock Fragments in 10 µl TE Buffer</li> |
| - | + | <li>Use 1 µl of each gBlock Fragment for a PCR with the Q5 Polymerase</li> | |
| - | <li> | + | <li>Perform the PCR Reaktion with the Primers Prefix and Suffix and an annealing temperature of 55 °C <br> (30 cycles)</li> |
| - | + | <li>Take 5 µl of the 50 µl reaction for DNA-gelelectrophoreses</li> | |
| + | <li>Restrict the other 45 µl with the restrictionenzymes EcoRI and PstI (10 U each) for 1 h at 37 °C</li> | ||
| + | <li>Ligate 5 µl of the reaction mix with 50 ng EcoRI/PstI resticted and purified pSB1C3 over night at 16 °C <br> For Ligation use the T4-Ligase and fresh T4-Ligase Buffer with ATP</li> | ||
| + | <li></li> | ||
| + | <li></li> | ||
| + | <li></li> | ||
| + | <li></li> | ||
| + | <li></li> | ||
</ul> | </ul> | ||
| + | <Br><Br><Br><Br><Br><Br><Br><Br> | ||
<font size="3" color="#F0F8FF" face="Arial regular"> | <font size="3" color="#F0F8FF" face="Arial regular"> | ||
<p text-aligne:left style="margin-left:50px; margin-right:50px"> | <p text-aligne:left style="margin-left:50px; margin-right:50px"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Construction of pSB1C3-petZ | Construction of pSB1C3-petZ | ||
<br><br> | <br><br> | ||
| - | <img alt="Zeta_toxin_pcr" src="/wiki/images/c/c7/Zeta_toxin_pcr_-_psb1C3_-_psb1c3_restiction.jpg" width="200" height="200" align="right"></a> | + | <img alt="Zeta_toxin_pcr" src="/wiki/images/c/c7/Zeta_toxin_pcr_-_psb1C3_-_psb1c3_restiction.jpg" width="200" height="200" align="right"style="margin-left:50px"; "margin-right:50px"></a> |
Revision as of 11:34, 29 September 2013
Labjournal
Assembly of the gBlocks
Based on the pDawn Plasmid we desinged the light induced kill switch and synthesized the construct as gBlock Fragments on IDT. We assembeled the 10 Fragments follow the Protocol:
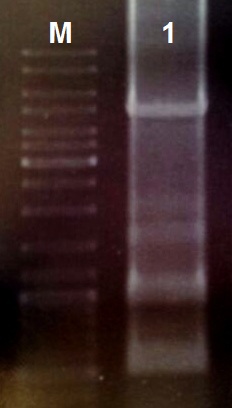
- Reconstitute the gBlock Fragments in 10 µl TE Buffer
- Use 1 µl of each gBlock Fragment for a PCR with the Q5 Polymerase
- Perform the PCR Reaktion with the Primers Prefix and Suffix and an annealing temperature of 55 °C
(30 cycles) - Take 5 µl of the 50 µl reaction for DNA-gelelectrophoreses
- Restrict the other 45 µl with the restrictionenzymes EcoRI and PstI (10 U each) for 1 h at 37 °C
- Ligate 5 µl of the reaction mix with 50 ng EcoRI/PstI resticted and purified pSB1C3 over night at 16 °C
For Ligation use the T4-Ligase and fresh T4-Ligase Buffer with ATP
Construction of pSB1C3-petZ
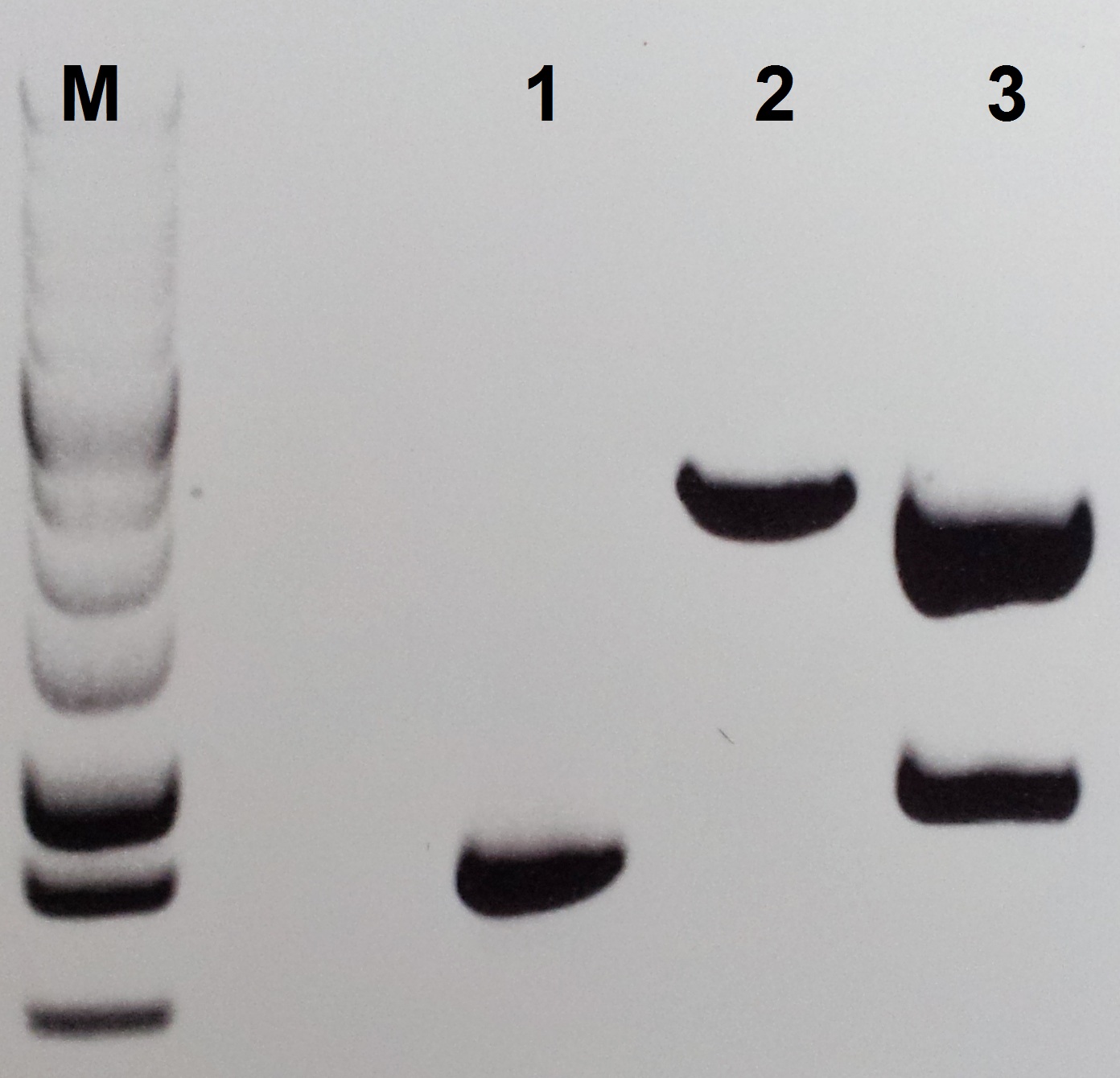
 "
"