Team:TU Darmstadt/safety/Labjournal
From 2013.igem.org
(Difference between revisions)
| Line 122: | Line 122: | ||
color: white; | color: white; | ||
} | } | ||
| + | |||
| + | |||
| + | |||
| + | dl.igemTUD2013gelpicture | ||
| + | { | ||
| + | border: 1px solid #000; | ||
| + | background-color: #109f71; | ||
| + | width: 120px; | ||
| + | text-align: center; | ||
| + | padding: 10px 5px 5px 5px; | ||
| + | float: right; | ||
| + | margin: 0 0 1em 1em; | ||
| + | } | ||
| + | |||
| + | .igemTUD2013gelpicture dt | ||
| + | { | ||
| + | font-weight: bold; | ||
| + | background-color: #131210; | ||
| + | color: #959289; | ||
| + | padding: 0 0; | ||
| + | margin-bottom: 10px; | ||
| + | } | ||
| + | |||
| + | .igemTUD2013gelpicture dd img | ||
| + | { | ||
| + | border: 1px solid #000; | ||
| + | width: 100px; | ||
| + | height: 200px; | ||
| + | } | ||
| + | |||
| + | .igemTUD2013gelpicture dd | ||
| + | { | ||
| + | margin: 0; | ||
| + | padding: 5px 5px 5px 5px; | ||
| + | font-size: 100%; | ||
| + | } | ||
| + | |||
| + | |||
</style> | </style> | ||
| Line 174: | Line 212: | ||
<Br> | <Br> | ||
| + | |||
| + | <dl class="igemTUD2013gelpicture"> | ||
| + | <dd><img alt="Assembly PCR with 10 gBlocks" src="/wiki/images/0/03/Assembly_pcr.jpg" width="100" height="200" align="right"></dd> | ||
| + | <dd>Assembly PCR with 10 gBlocks</dd> | ||
| + | </dl> | ||
| - | |||
<ul style="margin-left:50px; margin-right:50px; text-align:justify; "> | <ul style="margin-left:50px; margin-right:50px; text-align:justify; "> | ||
Revision as of 14:01, 29 September 2013
Labjournal
gBlocks assembly
We designed the light induced kill switch based on the pDawn plasmid. We ordered 10 gBlock fragments coding for our plasmid from IDT. We assembeld the fragments according to the following protocol:
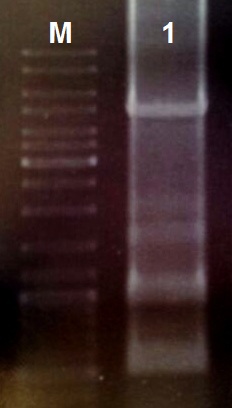
- Assembly PCR with 10 gBlocks
- Reconstitute the gBlock fragments in 10 µl TE buffer
- Use 1 µl of each gBlock fragment for a PCR with the Q5 Polymerase
- Perform the PCR reaction with primers coding for the prefix and suffix and an annealing temperature of 55 °C (30 cycles)
- Load an 0.8% agarose gel with 5 µl of PCR reaction (total volume 50 µl) and perform a DNA gel electrophoresis
- Cut the other 45 µl with the restriction enzymes EcoRI and PstI (10 U each) for 1 h at 37 °C
- Ligate 5 µl of the reaction mix with 50 ng EcoRI/PstI restricted and purified pSB1C3 over night at 16 °C
For Ligation use the T4-Ligase and fresh T4-Ligase Buffer with ATP - After heat-inactivation of the ligase by 80 °C for 15 min use 2 µl of the mix for heat-shock transformation
- Plate out the transformation mix on black petridishies with LB-cmp media
Construction of pSB1C3-pezT
For the construction of pSB1C3-petZ we isolated the pezT gene via PCR from the gBlocks C1 and C3 following this protocol:
- Use the primer pair Pre-PezT/For-PezT and an annealing temperature of 55 °C. For the isolation PCR follow the Q5 PCR protocol from NEB Biolabs.
- Use 5 µl for the control gel electrophoresis (1.2% agarose) and clean the rest of the reaction with the Wizard SV Gel and PCR Clean-Up System from Promega.
- Restrict 500 ng of the PCR product and 1 µg of pSB1C3 with EcoRI and PstI (10 U each) at 37 °C for 30 min.
- Perform the ligation with an molar ratio of 3:1 (insert:vector).
For Ligation use the T4-Ligase and fresh T4-Ligase Buffer with ATP - Transform 2 µl of the ligation mix into E. coli DH5α.
- Plate out the transformation on petri dishes with LB-cmp media.
 "
"