Team:TU-Munich/Notebook/Labjournal
From 2013.igem.org
| Line 195: | Line 195: | ||
[[File:TUM13_20130423_RFP_Generator_RFC25_AgeI_NgoMIV.png|500px]] | [[File:TUM13_20130423_RFP_Generator_RFC25_AgeI_NgoMIV.png|500px]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| Line 215: | Line 209: | ||
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer). | Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer). | ||
| + | </div> | ||
| + | |||
| + | =='''Monday, April 22nd'''== | ||
| + | |||
| + | <div class="safety_mechanism"> | ||
| + | |||
| + | === Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === | ||
| + | |||
| + | '''Investigator: Jeff, Leonie, Florian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3). | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="safety_mechanism"> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10 === | ||
| + | |||
| + | '''Investigator: Jeff, Leonie, Florian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Batch for analytical digestion for P7 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P7 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion for P8 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P8 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion for P9 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P9 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion for P10 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P10 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Incubation for 90 min at 37 °C. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 60 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |1 kbp ladder DNA ladder | ||
| + | |'''P7''' | ||
| + | |'''P8''' | ||
| + | |'''P9''' | ||
| + | |'''P10''' | ||
| + | |- | ||
| + | | | ||
| + | |'''Part is correct''' | ||
| + | |'''Part is correct''' | ||
| + | |'''Part is correct''' | ||
| + | |'''Part is correct''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[File:TUM13_20130423_RFP_Generator_RFC25_AgeI_NgoMIV.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | |||
| + | <!--- this closes the labbook--> | ||
</div> | </div> | ||
Revision as of 13:15, 24 April 2013

Labjournal
Week 1
Monday, April 22nd
Transformation of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Transformation of Phytochrome B for protein fusion.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Sequencing of RFP-Generator (RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Sequencing of RFP-Generator (RFC25, pSB1C3)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Tuesday, April 23rd
Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Procedure:
- pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5).
Procedure:
- Batch for analytical digestion for P4 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P4 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P5 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P5 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P4 | P5 |
| Mutation successful | Mutation successful! |
- Parts are compliant and do not contain RFC25 forbidden restriction sites.
Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
Monday, April 22nd
Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
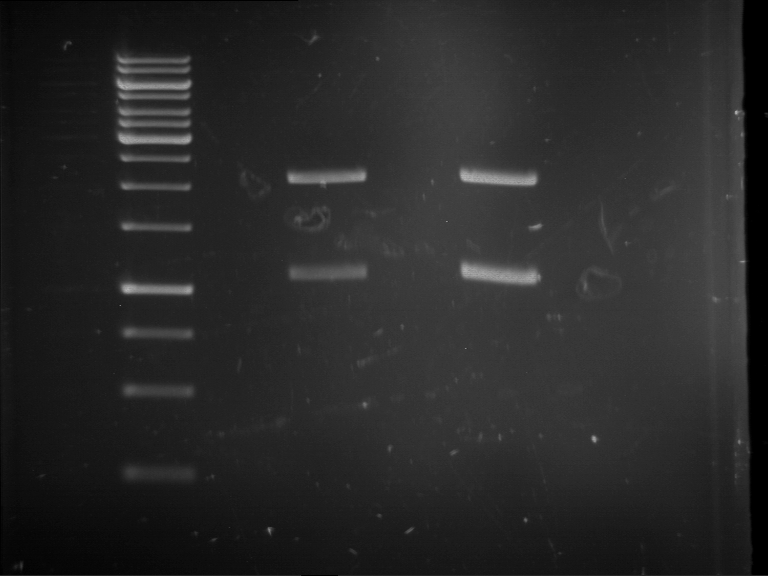
Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10.
Procedure:
- Batch for analytical digestion for P7 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P7 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P8 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P8 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P9 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P9 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P10 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P10 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P7 | P8 | P9 | P10 |
| Part is correct | Part is correct | Part is correct | Part is correct |
 "
"